Fast Kernel Methods Le Song Machine Learning II: Advanced Topics
advertisement

Fast Kernel Methods
Le Song
Machine Learning II: Advanced Topics
CSE 8803ML, Spring 2012
Weight space view of GP
Assume linear regression model
𝑓 𝑥 𝑤 = 𝑥⊤𝑤
𝑦 =𝑓+𝜖
𝜖 ∼ 𝒩 0, 𝜎 2
Let 𝑌 =, 𝑦1 , … 𝑦𝑛 , 𝑋 = 𝑥1 , … , 𝑥𝑛 , Likelihood of observations
𝑃( 𝑦𝑖
𝑛
𝑖=1
𝑥𝑖
𝑛
𝑖=1 , 𝑤
= 𝒩 𝑋 ⊤ 𝑤, 𝜎 2 𝐼
Assume a Gaussian prior over parameters
𝑃 𝑤 = 𝒩(0, 𝐼)
Apply Bayes’ theorem to obtain posterior
𝑃 𝑤 𝑌, 𝑋 ∝ 𝑃 𝑌 𝑋, 𝑤 𝑃(𝑤)
Matrix inversion
lemma connects
two views of GP
2
Gaussian processes
An infinite collection of Gaussian random variables
Indexed by covariate 𝑥, the set of index can be infinite, hence an
infinite collection of Gaussian random variables.
Mean function 𝑚(𝑥)
Covariance functions 𝑘(𝑥, 𝑥’) of two Gaussians indexed by 𝑥 and 𝑥’
Covariance
𝑘 𝑥𝑖 , 𝑥𝑗
To generate a sample from GP
Gaussian variable 𝑓𝑖 , 𝑓𝑗 are indexed by
𝑥𝑖 , 𝑥𝑗 respectively, and their covariance
(𝑖𝑗-th entry in Σ) defined by 𝑘 𝑥𝑖 , 𝑥𝑗
𝑓𝑖
𝑓𝑗
Generate 𝑁 iid. samples: 𝑦 =
𝑦1 , … , 𝑦𝑁 ⊤ ∼ 𝒩 0; Σ
𝑥𝑖
𝑥𝑗
3
Covariance function of Gaussian processes
For any finite collection of indices 𝑥1 , 𝑥2 , … , 𝑥𝑛 , the covariance
matrix is positive semidefinite
Σ=𝐾=
𝑘 𝑥1 , 𝑥1
𝑘 𝑥2 , 𝑥1
𝑘 𝑥1 , 𝑥2
𝑘 𝑥2 , 𝑥2
⋮ ⋮
𝑘(𝑥𝑛 , 𝑥1 ) 𝑘(𝑥𝑛 , 𝑥2 )
⋯ 𝑘 𝑥1 , 𝑥𝑛
⋯ 𝑘 𝑥2 , 𝑥𝑛
⋱ ⋮
⋯ 𝑘(𝑥𝑛 , 𝑥𝑛 )
The covariance function needs to be a kernel function over the
indices!
Eg. Gaussian RBF kernel
𝑘 𝑥, 𝑥 ′ = exp −
1
2
𝑥 − 𝑥′
2
4
Using Gaussian process for nonlinear regression
Observing a dataset 𝐷 =
𝑥𝑖 , 𝑦𝑖
𝑛
𝑖=1
Prior 𝑃(𝑓) is Gaussian process, like a multivariate Gaussian,
therefore, posterior of 𝑓 is also a Gaussian process
Bayesian rule 𝑃 𝑓 𝐷 =
𝑃 𝐷 𝑓 𝑃(𝑓)
𝑃(𝐷)
Everything else about GPs follows the basic rules of
probabilities applied to multivariate Gaussians
5
Parameter tuning in GP
We want to select features or tune hyperparameters
For instance, covariance function
𝑘 𝑥𝑖 , 𝑥𝑗 = 𝑣0 exp −
𝐷
𝑑=1
𝑥𝑖𝑑 −𝑥𝑗𝑑
𝛼
+ 𝑣1 + 𝑣2 𝛿𝑖𝑗
𝑟𝑑
𝑟𝑑 can be used for feature selection
𝛼 𝑎𝑛𝑑 𝑣 can be related to other property of Gaussian process
Use marginal likelihood ln 𝑃( 𝑦𝑖
do feature selection
𝑛
𝑖=1
𝑥𝑖
𝑛
𝑖=1
to tune 𝑟𝑑 , 𝛼 𝑎𝑛𝑑 𝑣 to
6
GP for classification
Regression model
𝑓|~𝐺𝑃 𝑚 𝑥 , 𝑘 𝑥, 𝑥 ′
2
𝑦|𝑥, 𝑓 ∼ 𝒩 𝑓(𝑥), 𝜎𝑛𝑜𝑖𝑠𝑒
𝐼
Classification model
Give data
𝑥𝑖 , 𝑦𝑖
𝑓|~𝐺𝑃 𝑚 𝑥 , 𝑘
𝑛
𝑖=1
𝑥, 𝑥 ′
where 𝑦𝑖 ∈ {−1, +1}
𝑦|𝑥, 𝑓 ∼ 𝑝 𝑦|𝑓(𝑥)
7
Relate GP to class probability
Transform the continuous output of Gaussian Process to a
value between [-1,1] or [0,1]
With binary outputs, the joint distribution of all variables in the
model is no longer Gaussians
The likelihood is also not Gaussian, so we will need to use
approximate inference to compute the posterior GP (Laplace
approximation, sampling)
8
Connection to kernel support vector machines
1
𝑤 2
min
𝑤
2
+𝐶
𝑗 𝜉𝑗
𝑠. 𝑡. 𝑤 ⊤ 𝜙(𝑥𝑗 ) + 𝑏 𝑦𝑗 ≥ 1 − 𝜉𝑗 , 𝜉𝑗 ≥ 0, ∀𝑗
𝜉𝑗 : Slack variables
Can be equivalently written as
a hinge loss function
1 − 𝑦𝑖 (𝑤 ⊤ 𝜙(𝑥𝑖 )) +
= 1 − 𝑦𝑖 𝑓𝑖 +
9
Connection to kernel support vector machines
We can rewrite the kernelized SVM as:
1
2
min 𝑓 ⊤ 𝐾 −1 𝑓 + 𝐶
𝑓
𝑖
1 − 𝑦𝑖 𝑓𝑖
+
We can write the negative log of a GP likelihood as
1
2
min 𝑓 ⊤ 𝐾 −1 𝑓 +
𝑓
𝑖 ln 𝑝(𝑦𝑖 |𝑓𝑖 ) +
𝑐
With Gaussian process we
Handle uncerntainty in unknown function 𝑓 by averaging, not
minimization
Can learn the kernel parameters and features using marginal
likelihood
Can incorporate interpretable noise models and priors, can
sample
10
Computation issue of GP and kernel methods
𝑓 𝑥 | 𝑥𝑖 , 𝑦𝑖
𝑛
𝑖=1
~𝐺𝑃 𝑚𝑝𝑜𝑠𝑡 𝑥 , 𝑘𝑝𝑜𝑠𝑡 𝑥, 𝑥 ′
2
𝑚𝑝𝑜𝑠𝑡 𝑥 = 𝑘 𝑥, 𝑋 𝐾 + 𝜎𝑛𝑜𝑖𝑠𝑒
𝐼
𝑘𝑝𝑜𝑠𝑡
𝑥, 𝑥 ′
=
𝑘(𝑥, 𝑥 ′ )
−1 ⊤
𝑌
− 𝑘 𝑥, 𝑋 𝐾
−1
2
+ 𝜎𝑛𝑜𝑖𝑠𝑒 𝐼 𝑘
𝑥′, 𝑋
⊤
Inverting kernel matrix 𝐾 is computationally intensive
Suppose there are 𝑛 data points, the computation cost is 𝑂 𝑛3
Need to reduce the computational cost by some type of
approximation
𝑅
𝐾
≈
𝑅⊤
11
Kernel low rank approximation
Incomplete Cholesky factorization of kernel matrix 𝐾 of size
𝑛 × 𝑛 to 𝑅 of size 𝑑 × 𝑛, and 𝑑 ≪ 𝑛
𝑅
≈
𝐾
𝑅⊤
𝑅
≈
𝐴
𝑓 𝑥 | 𝑥𝑖 , 𝑦𝑖
𝑛
𝑖=1
𝑅⊤
~𝐺𝑃 𝑚𝑝𝑜𝑠𝑡 𝑥 , 𝑘𝑝𝑜𝑠𝑡 𝑥, 𝑥 ′
2
𝑚𝑝𝑜𝑠𝑡 𝑥 = 𝑅𝑥⊤ 𝑅𝑅⊤ + 𝜎𝑛𝑜𝑖𝑠𝑒
𝐼
𝑘𝑝𝑜𝑠𝑡
𝑥, 𝑥 ′
= 𝑅𝑥𝑥 −
𝑅𝑥⊤
𝑅𝑅⊤
−1
+
𝑅𝑌 ⊤
−1
2
𝜎𝑛𝑜𝑖𝑠𝑒 𝐼 (𝑅𝑅⊤ )𝑅𝑥
12
Incomplete Cholesky Decomposition
We have a few things to understand
Gram-Schmidt orthogonalization
Given a set of vectors V = {𝑣1 , 𝑣2 , … , 𝑣𝑛 }, find a set of orthonormal
basis 𝑄 = 𝑢1 , 𝑢2 , … 𝑢𝑛 , 𝑢𝑖⊤ 𝑢𝑗 = 0, 𝑢𝑖⊤ 𝑢𝑖 = 0
QR decomposition
Given a set of orthonormal basis 𝑄, compute the projection of 𝑉
onto 𝑄, 𝑣𝑖 = 𝑗 𝑟𝑗𝑖 𝑢𝑗 , 𝑅 = 𝑟𝑗𝑖
𝑉 = 𝑄𝑅
Cholesky decomposition with pivots
𝑉 ≈ 𝑄 : , 1: 𝑘 𝑅 1: 𝑘, ∶
Kernelization
𝑉 ⊤ 𝑉 = 𝑅⊤ 𝑄 ⊤ 𝑄𝑅 = 𝑅⊤ 𝑅 ≈ 𝑅 1: 𝑘, ∶
𝐾 = Φ⊤ Φ ≈ 𝑅 1: 𝑘, ∶
⊤
⊤
𝑅 1: 𝑘, ∶
𝑅 1: 𝑘, ∶
13
Gram-Schmidt orthogonalization
Given a set of vectors V = {𝑣1 , 𝑣2 , … , 𝑣𝑛 }, find a set of orthonormal basis
𝑄 = 𝑢1 , 𝑢2 , … 𝑢𝑛 , 𝑢𝑖⊤ 𝑢𝑗 = 0, 𝑢𝑖⊤ 𝑢𝑖 = 0
𝑢1 can be found by picking an arbitrary 𝑣1 and normalize
𝑢1 =
𝑣1
𝑣1
𝑢2 can be found by picking a vector 𝑣2 and subtract out
multiple of 𝑢1 , and then normalize
𝑎2 = 𝑣2 − < 𝑣2 , 𝑢1 > 𝑢1
𝑢2 =
𝑎2
𝑎2
𝑣2
𝑎2
𝑢2
𝑣1
𝑎𝑖 = 𝑣𝑖 −
𝑖−1
𝑗=1
< 𝑣𝑖 , 𝑢𝑗 > 𝑢𝑗
𝑢1
14
Orthonormal basis
First, every 𝑢 is normalized to unit norm
𝑢⊤ 𝑢 = 1
Two 𝑢𝑖 𝑎𝑛𝑑 𝑢𝑗 are orthogonal to each other
𝑢𝑖⊤ 𝑢𝑗 = 0
Eg. 𝑢1⊤ 𝑢2 ∝ 𝑢1⊤ 𝑣2 − < 𝑣2 , 𝑢1 > 𝑢1
=< 𝑣2 , 𝑢1 >−< 𝑣2 , 𝑢1 >< 𝑢1 , 𝑢1 > = 0
More generally, prove by induction
All previous ones are orthonormal
Show the new one is orthogonal to all previous ones
15
QR decomposition
Essentially Gram-Schmidt orthogonalization, keep both the
orthonormal basis and weight of the projection
Given a set of vectors V = {𝑣1 , 𝑣2 , … , 𝑣𝑛 }, find a set of
orthonormal basis 𝑄 = 𝑢1 , 𝑢2 , … 𝑢𝑛 using Gram-Schmidt
orthogonalization
The projection of 𝑣𝑖 on to basis vector 𝑢𝑗 is 𝑟𝑗𝑖 =< 𝑣𝑖 , 𝑢𝑗 >
𝑣1 = 𝑢1 < 𝑢1 , 𝑣1 >
𝑣2 = 𝑢1 < 𝑢1 , 𝑣2 > +𝑢2 < 𝑢2 , 𝑣2 >
𝑣3 = 𝑢1 < 𝑢1 , 𝑣3 > +𝑢2 < 𝑢2 , 𝑣3 > +𝑢3 < 𝑢3 , 𝑣3 >
…
𝑣𝑖 =
𝑖
𝑗=1
< 𝑣𝑖 , 𝑢𝑗 > 𝑢𝑗
16
QR decomposition
Because use the original data point to form basis vectors,
vector 𝑣𝑖 only have 𝑖 nonzeros components
𝑣𝑖 =
𝑖
𝑗=1
< 𝑣𝑖 , 𝑢𝑗 > 𝑢𝑗 =
𝑖
𝑗=1 𝑟𝑗𝑖 𝑢𝑗
Collect terms into matrix format
𝑉 = 𝑣1 , … , 𝑣𝑛 , 𝑣𝑖 ∈ 𝑅𝑑
𝑄 = (𝑢1 , … , 𝑢𝑑 )
zeros
𝑅 = (𝑟:𝑖 , … , 𝑢:𝑛 )
17
QR decomposition with pivots
QR decomposition
𝑉=
𝑄 = (𝑢1 , … , 𝑢𝑑 )
zeros
𝑅 = (𝑟:𝑖 , … , 𝑢:𝑛 )
If we only choose a few basis vectors, then its approximation
The basis vectors is formed from the original data points
how to order/choose from the original data points?
such that small approximation error?
order/choose from data points: choosing pivots
18
Cholesky decomposition
𝐾 is symmetric and positive definite matrix, then 𝐾 can be
decomposed as
𝐾 = 𝑅⊤ 𝑅
Since 𝐾 is a kernel matrix, we can find an implicit feature space
𝐾 = Φ⊤ Φ, where Φ = 𝜙 𝑥1 , … , 𝜙 𝑥𝑛
QR decomposition on Φ = QR
𝐾 = R⊤ Q⊤ QR = 𝑅⊤ 𝑅
Incomplete Cholesky decomposition
Use QR decomposition with pivots
𝐾 ≈ 𝑅 1: 𝑑, : ⊤ 𝑅(1: 𝑑, : )
𝑅
𝐾
≈ 𝑅⊤
19
Incomplete Cholesky Decomposition
Key question I: how to choose pivots?
Greedy approach: choose the next pivot with the largest norm
after projecting out components on previous basis vectors
𝑅
𝐾
≈
𝑅⊤
Key question II: do we need to form the full kernel matrix 𝐾 in
order to compute the approximation?
Can we working directly with data point and kernel function?
Can we make the computation linear in the number of data
pioonts?
20
Incomplete Cholesky decomposition: Matlab
Kernel entries can
be computed on
the fly
Computation
𝑂 𝑛𝑑2 number
of kernel
evaluation
21
Random features for kernels
Incomplete Cholesky decomposition essentially approximate
an infinite dimensional feature space with a small number of
chosen basis vector
Is there a simpler and even faster way to choose the basis
vectors?
Random features use randomly chosen basis vector to
approximate the feature space!
What are the basis vectors?
What type of randomness to use?
22
Translational invariance kernel
Kernel value only depends on the difference between two data
points
𝑘 𝑥, 𝑦 = 𝑘 𝑥 − 𝑦 = 𝑘(Δ)
A translational invariance kernel 𝑘(Δ) is the Fourier transformation
of a non-negative measure (Bochner theorem)
Eg.
23
Random features
What basis to use?
′
𝑒 𝑗𝜔 (𝑥−𝑦) can be replaced by cos(𝜔 𝑥 − 𝑦 ) since both 𝑘 𝑥 − 𝑦
and 𝑝 𝜔 real functions
cos 𝜔 𝑥 − 𝑦 = cos 𝜔𝑥 cos 𝜔𝑦 + sin 𝜔𝑥 sin 𝜔𝑦
For each 𝜔, use feature [cos 𝜔𝑥 , sin 𝜔𝑥 ]
What randomness to use?
Randomly draw 𝜔 from 𝑝 𝜔
Eg. Gaussian RBF kernel, drawn from Gaussian
24
Random features: Matlab
Random features usually need more feature dimensions than
incomplete Cholesky decomposition
25
Random Features
MIST digit dataset
26
Nystrom’s method for kernel matrix
Use subblock of the kernel matrix to approximate the entire
kernel matrix G
27
String Kernels
Compare two sequences for similarity
K(
ACAAGAT
GCCATTG
TCCCCCG
GCCTCCT
GCTGCTG
,
GCATGAC
GCCATTG
ACCTGCT
GGTCCTA
)=0.7
Exact matching kernel
Counting all matching substrings
Flexible weighting scheme
Does not work well for noisy case
Successful applications in bio-informatics
Linear time algorithm using suffix trees
28
Exact matching string kernels
Bag of Characters
Count single characters, set 𝑤𝑠 = 0 for 𝑠 > 1
Bag of Words
s is bounded by whitespace
Limited range correlations
Set 𝑤𝑠 = 0 for all 𝑠 > 𝑛 given a fixed 𝑛
K-spectrum kernel
Account for matching substrings of length 𝑘, set 𝑤𝑠 = 0 for all
𝑠 ≠𝑘
29
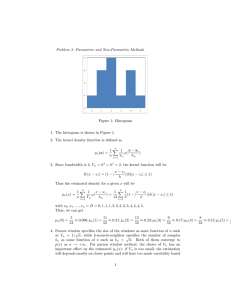
Suffix trees
Definitions: compact tree built from all the suffixes of a string.
Eg. suffix tree of ababc denoted by S(ababc)
Node Label = unique path from the root
Suffix links are used to speed up parsing of strings: if we are at
node 𝑎𝑥 then suffix links help us to jump to node 𝑥
Represent all the substrings of a given string
Can be constructed in linear time and stored in linear space
Each leaf corresponds to a unique suffix
Leaves on the subtree give number of occurrence
30