Thermodynamics of electron flow in the bacterial decaheme cytochrome MtrF
advertisement

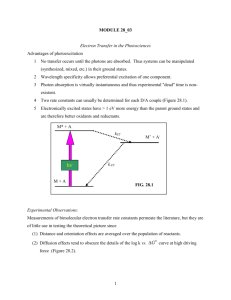
Thermodynamics of electron flow in the bacterial decaheme cytochrome MtrF Breuer M.(1), Zarzycki P. P.(2), Rosso K. M.(2), Blumberger J.(1)* (1) University College London, Gower Street, London WC1E 6BT, UK (2) Pacific Northwestern National Laboratory, 902 Battelle Boulevard, Richland, WA 99352, USA * Email: j.blumberger@ucl.ac.uk Abstract Very recently the first crystal structure of a bacterial decaheme ctype cytochrome has been resolved1. Shewanella oneidensis and other metal-respiring bacteria use such biological `wire’ proteins to shuttle electrons across the outer membrane on to extracellular solid metal oxides, e.g. Fe2O3, which serve as terminal electron acceptors. Investigation of the electron transport properties of these multi-heme proteins not only serves to promote understanding of biological charge transfer and transport processes but also holds out the prospect of promising bionanotechnological applications. The heme cofactors in the particular protein under investigation, MtrF, are arranged as a chain of four approximately coplanar heme groups separating two triple-stacks of approximately parallel heme cofactors, altogether constituting a ‘molecular wire’. In order to investigate the electron transport properties of MtrF, reduction potentials of individual heme cofactors have been calculated using classical molecular dynamic simulation combined with thermodynamic integration along two different paths. Results from these calculations are discussed and compared with each other as well as with experimental reduction potentials obtained from protein film voltammetry measurements. Finally, electronic coupling matrix element values for adjacent hemes as obtained using the fragment-orbital DFT method are presented. Background • Shewanella oneidensis strain MR-1: a dissimilatory metal-reducing bacterium • Capable of using extracellular solid metal oxides as terminal electron sinks • A complex system of many multiheme proteins transfers electrons to the cell exterior Electron transfer model • Marcus theory is used to model electron transfer from heme to heme4: • Rate constant of electron transfer kET depends on electronic coupling matrix element H12, reorganization energy λ and free energy of electron transfer ∆ A for a given temperature T • These quantities can be calculated using computational methods Reduction potentials • Free energy difference between two states can be computed via thermodynamic integration: where the potential energy U is a function of λ and the canonical average is taken over the corresponding potential energy surface • Classical molecular dynamics simulations were used to obtain reduction potentials via thermodynamic integration, scaling atomic charges linearly with λ: • MtrF: the first outer membrane decaheme c-type cytochrome whose crystal structure was determined • This yields: • Crystal structure enables new insights into biological charge transport processes • Two different pathways were used: Fig. 7: Free energy landscapes based on the reduction potentials obtained via the two thermodynamic integration pathways. Negative values from fig. 5 are used and shifted to 0.0 eV for heme 6; free energy barriers are approximated by assuming λ = 0.7 eV. Electronic couplings • Electronic coupling matrix element is defined as: between two diabatic states a and b • Fragment-orbital DFT (FO-DFT): Approximate H12 by i.e. as Kohn-Sham Hamiltonian matrix elements between HOMOs of donor and reduced acceptor; both are treated as isolated – Oxidation of a reduced heme to directly yield its reduction potential – Electron transfer between a reduced and an oxidized heme to yield electron transfer free energies (which in turn provide relative reduction potentials) • Assumption: Electron supply to protein is a limiting factor Fig. 2: Proposed electron transfer Fig. 1: Shewanella oneidensis strain MR-1.2 pathways in S. oneidensis from cytoplasmic membrane (CM) to outer membrane (OM) involving different cytochromes.3 --> Electron transport, not hole transport --> Treat all hemes but one as oxidized 5 MtrF: Structure Fig. 8: Electronic coupling matrix elements (absolute values) between adjacent hemes in units of mHa as calculated using FODFT. Each coupling was calculated on a structure minimized with both respective hemes half-reduced (and all others oxidized). 4 3 Conclusions 2 1 7 6 8 • Thermodynamic integrations along two different pathways have been carried out in order to calculate reduction potentials of individual hemes 9 10 Fig. 3: Crystal structure of MtrF. Heme groups are highlighted in colour.1 Fig. 5: Reduction potentials for individual hemes in eV as obtained via the two thermodynamic integration pathways. Blue: values from oxidation, green: from electron transfer. (The latter obtained from the transfer free energies using an arbitrary offset and shifting to best match values from oxidation pathway.) Red: differences. • MtrF: a c-type cytochrome containing ten heme cofactors • Both data sets show correlation with experimental data to some degree but disagree with each other • No conclusions yet regarding possible thermodynamic preferences regarding direction of electron flow • Electronic coupling matrix elements between hemes have been calculated using FO-DFT, showing variations over two orders of magnitude References • Two of four domains contain α -helices, the other containβ -sheets; many random coils present [1] Clarke T.A. et al. PNAS 2011, published ahead of print May 23, 2011, doi:10.1073/pnas.1017200108. [2] Image source: http://idw-online.de/pages/de/news370736 (16/05/2011). [3] Coursolle D. and Grainick J. A. Mol. Microbiol., 2010, 77, 995. [4] Marcus R. A. J. Chem. Phys., 1965, 43, 679. • Each heme is covalently bound by two cysteines (CXXCH binding motif) and coordinated by two histidines Fig. 3: Individual heme cofactor including covalently bound cysteine and coordinated histidine (both truncated). Green: iron; blue: nitrogen; light blue: carbon; red: oxygen; yellow: sulphur; white: hydrogen. • The first crystal structure of a bacterial decaheme c-type cytochrome enables new insights into biological charge transfer processes Acknowledgements Fig. 6: Comparison of individual heme reduction potentials and experimental values (protein film voltammetry, PFV). PFV values are relative to the standard hydrogen electrode; values from thermodynamic integration are shifted (values from each method individually) to the best possible match to PFV values. M. Breuer gratefully acknowledges a joint studentship sponsored by University College London and the Pacific Northwestern National Laboratory. The work in this study has been carried supercomputers Hector, Chinook and Legion. out on the