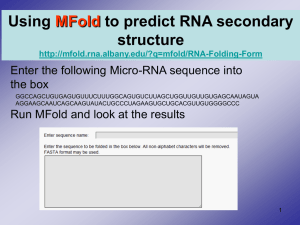
From: ISMB-95 Proceedings. Copyright © 1995, AAAI (www.aaai.org). All rights reserved.
MMDB:An ASN.1 Specification
for Macromolecular Structure
Hitomi Ohkawa,JamesOstell and StephenBryant
National Center for Biotechnology Information
National Library of Medicine, National Institute of Health, Bldg.38A, Rm.SN805
8600 Rockville Pike, Bethesda, MD20894 USA
ohkawa@ncbi.nlm.nih.gov,ostell@ncbi.nlm.nih.gov, bryant@nebi.nlm.nih.gov
Abstract
Wepresent an exchangespecification for data describing the
three-dimensionalstructure of biological macromolecules.
The
specification was designed for MMDB,
a Molecular Modeling
Database supported by the National Center for Biotechnology
Information(NCBI),basedon informationfromthe Protein Data
Bank(PDB).In the MMDB
specification, the chemicalstructures
of moleculesare describedhierarchicallyas connectivitygraphs,
to directly support comparisonby subgraph isomorphismor
assignmentalgorithms.Three-dimensional
coordinatesare linked
unambiguously
to nodesin the chemicalgraph, so that homologyderived structures maybe generateddirectly fromalignmentof
chemicallysimilar groups.In conversionto this form,data from
PDBare extensivelyvalidated, so as to providea descriptionof
chemicaland spatial structure that is as accurate as possible.
Thesechangesin formatand content of the known
structure data
are intended to support developmentof intelligent molecular
modelingapplications that makeuse of this invaluableinformation resource.
Description of MMDB
Wepresent a data exchangespecification for information
describingthe three-dimensionalstructure of biological macromolecules. The specification was designed for MMDB,
a
MolecularModelingDatabase supported by the National Center for BiotechnologyInformation,NCBI. MMDB
is based on
informationfromthe Protein DataBank(Bernstein et al. 1977),
modified in form and content to produce a macromolecular
structure database readily usable by computationalbiologists
and developersof molecularmodelingsoftware.
The MMDB
specification is written in ASN.I, an ISOOpen
SystemsInterconnection Standard used for formal, standardized data exchangeabovethe level of specific software and
hardware (Rose 1990, Ostell et al. 1994). Macromolecular
structure data in this form maybe read into computermemory
using a suite of softwaretools also available fromNCBI,in the
formof C-languagesubroutinelibraries (Ostell et al. 1994).
This softwareautomaticallytranslates an ASN.I stream into C
data structureswhichare fully atomic,in the sensethat all parsable data items from PDBare represented as individual
numericor character values. Softwaredevelopersmaytlaerefore directly retrieve and manipulatedata items relevant to
molecularmodelingby C subroutinecall, instead of by parsing PDBtext files. TheC data structure declarations ate producedautomatically from the ASN.1specification, and data
item namesand semantics are described fully by the MMDB
specificationpresentedhere.
Molecularmodelinginvolves comparisonof the chemical
structures of twomoleculesto producean atom-by-atom
mapping, fromwhichthe partial spatial structure of one molecule
maybe inferred from that of the other. The information
required is an unambiguous
description of chemicalstmetu~
in the formof a chemicalgraph, and an unambiguous
linking
of spatial coordinatedata to atomsformingthe nodesof this
graph. To facilitate molecularmodelingMMDB
therefore provides this information explicitly. Software maydirectly
retrieve the data items neededfor sequencealignmentor subgraph isomorphismcalculations, and need not encode the
complexlogic required to deduce covalent structure from
atomand residue namesand other conventions employedby
PDB.Homology
modelsderived in this waymayalso be representedexplicitly.
Chemicalgraphs in MMDB
are represented in a fashion
similar to that proposedby the ChemicalAbstractsServicein
the CXFspecification (Mockus&Steckert 1994, Moeims
Steckert1995)and by the International Unionof Crystallography in their mmCIF
specification (Shindyalovet al. 1994,
Shindyalov et al. 1995, Wodaket al. 1994). Biomolecular
assemblies are organized as a chemicalhierarchy of atoms,
residues, molecules,with subgraphsfor biopolymerresidues
givenby referenceto a standarddictionary. Thestandardsubgraph dictionary distributed with MMDB
includes the 20
aminoacids naturally occurringin proteins and the 8 ribonucleotide and deoxyribonucleotide groups occurring in RNA
and DNA.Construction of MMDB
requires validation of PDB
data against this dictionary, andtherefore identifies a number
Ohkawa
259
of inconsistenciesand errors as occurrencesof nonstandardresidue groups. Subgraphsfor these and true non-polymercomponents suchas protein cofactors are constructedby reference to
any explicit connectivitydata providedby PDB,withvalidation
by stereochemicalcalculations basedon atomiccoordinates.
Atomiccoordinate data in MMDB
retain all information
providedby PDB,includingcrystallographic modelswith alternate conformationsresulting from statistical disorder, and
NMR-derived
modelsrepresented as an ensembleof alternative
structures. Wehave attempted to represent this information
unambiguously,
a process requiring considerable validation of
any multiple-coordinate data provided by PDB.For manycomputational biologyapplications, however,it is useful to havea
simplifiedmodelin whichonlya single "best" coordinateis provided for each atom in the chemicalgraph. To this end MMDB
provides a single-coordinate-per-atom modelas producedby
the PK.Banalysis suite (Bryant 1989), a "view"of macromolecular structure whichhas been tested in manyapplications.
MMDB
also provides a further simplified single-coordinateper-residueview, intendedfor graphical applications and rapid
networktransmission.
MMDB
also allows for non-atomicrepresentations of structare, suchas densityor surfacemodels.Theseare not presentin
PDB,but the correspondingobject types maynonetheless prove
useful to computationalbiologists whoencounter these commonrepresentations. Structural features are defined in MMDB
as generic descriptors and sets of properties to bd associated
with atomsor residues, or a region in space. Thisdefinition is
sufficient to represent secondarystructure andsite annotations
as provided by PDBor proposed in mmCIF,but also general
enough to accommodatenew data. One might, for example,
describe the electrostatic potential at points on a surface grid
defined in the space of an atomicmodelfrom crystallography.
Onemightsimilarly describe the local environmentcategories
to be associated with a set of residues. Theseobject types in
MMDB
are intended primarily to facilitate developmentof new
applications.
Figure 1 shows a diagrammatic representation of the
MMDB
specification, giving an overview of relationships
amongdata items and the design concepts behind MMDB.
Appendix1 lists the completeMMDB
specification and constitutes the bodyof this paper. Thespecification itself includes
detailed commentswhichexplain data item semantics and the
mannerin which data items from PDBare mappedinto MMDB.
Thespecification, corresponding
C structure definitions, and I/
O routines are available via anonymous ftp from
ncbi.nlm.nih.gov. ExampleC programs are also provided,
including one that produces from MMDB
a validated, PDB-for-
260
ISMB-95
mattedfile. MMDB
data files are available for ftp, but mayalso
be accessedvia client software addressing the Entrez server
(NCBI1994), whichwill provide in ASN.1form damdescribing the three-dimensionalstructure of maeromolecules,
as well
as their sequences,
andcitations to relevantscientific literature.
References
Benson,D.A.; Bognski,M.; Lipman,DJ.; and Ostell, J. 1994.
Genbank.Nucleic Acids Research22:3441-3444.
Berstein,F.C.; Koetzle,T.F.; Williams,G.J.B.; Meyer,E.F.;
Brice, M.D.;Rodgers,J.R.; Kennerd,04 Simanouchi,T.; and
Tasumi, M. 1977. The Protein Data Bank:A computer-based
archival file for macromolecular
structures. Journalof Molecular Biology112:535-542.
Bryant, S.H. 1989. PKB:A ProgramSystemand Data Base for
Analysisof Protein Structure. Proteins5:233-247.
Mockus,J., and Steckert, T.D. 1994. ChemicalexchangeFormatVersion1.0. ChemicalAbstractsService, A Divisionof the
AmericanChemicalSociety.
Mockus,J., and Steekert, "I.D. 1995. CXF- the Chemical
eXchangeFormat. Gragg, C.E., and Mockns,J. eds. Chem/c~d
DataStandards:Databases,DataInterchange,andlnformation
Systems - 2 vol., ASTM
STP1298. Philadelphia, PA: American
Societyfor Testingand Materials.
NCBI.1993. Entrez Release 6.0. Bethesda, MD:NCBI,NLM/
NIH.
Ostell, J. 1994. NCBISoftwareDevelopment
Toolkit Version
1.9. NCBI,NLM/NIH,
Bethesda, MD.
Rose, M.T.1990. The OpenBook(A Practical Perspective on
OSI). Englewood
Cliffs, N J: PrenticeHall.
Shindyalov,I.N.; Chang,W;Pu, C; and Bourne,P.E. 1994. Macromolecular query language (MMQL):
prototype data model
and implementation.Protein Engineering7(11): 1311-1322.
Shindyaiov,L.N.; Chang,W;Cooper,J.A; and Bourne,P.E.
1995. Designand Useof a SoftwareFramework
to ObtainIrfformarionDerivedfrom Macromolecular
Structure Data. In Proceedingsof the Twenty-Eighth
AnnualHawaiiInternational
Conferenceon SystemSciences, 207-216.Hawaii:The Institute
of Electrical and ElectronicsEngineers,Inc.
Wodak,S. 1994. Proceedingsof the EuropeanMacro-Molecular
CIF Workshop.Brussels.
Q-O
"5 o
II
°~
J ..........
mE
"""~ e"
j
L
6
-
~g ~e
Em
E
mE
-~°~
Q.
Q}
r-
t
~ ~-~
,tl
Ohkawa
261
262
ISMB-95
J
J
~I
~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~
~.~~~
,~
.
.~
-~
-g~
~ ~ -
~ ~ ~ ~ ~ ~ U ~ " ~ ~ ~ ~ ~ ~ ~ ~
~~-~_~-~_~
~-
J
|
!
~ ,
.Jl
~
~
,~
~i~ ~ !!
|~
~
ill,
l
~ ~,.~ ~
Ohkawa
263
I
.++’+
II!
264
ISMB--95
Ohkawa
265
|
8
|
z
.~.~
!!! ,,
.!t
m
!i,
!J
~,~
J
!
0
o ,~ ~
i
-
!! i~l
-!
~i
¯E ’
11
O
tt
1
266 ISMB--95
.I
.| a
ii
|
I
o~
t
t
i
i
.O
U
|
1
!
i
i
,H
I
,ll
|
!!!
!!!
t
15
t
1
|
x
|
’s
1,ll
.g
E
Z
°i!
i
ii -i
jJ
t
! ~11 i
t
!
I
~
~I
|
| ~’
,.,
I
aa
l
g
,,B
J
|
J
.i
11
m
~]~i
II
|
J
i’
!,J
j
SJ
I
|
!
’i
I~
t_ i
..,..
I l,,Iii
Ohkawa
267