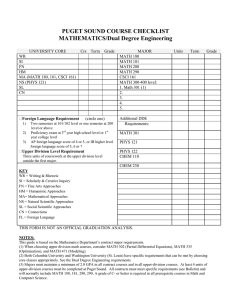
New developments in enhanced conformational sampling using molecular dynamics

New developments in enhanced conformational sampling using molecular dynamics
Mark E. Tuckerman
Dept. of Chemistry and Courant Institute of Mathematical Sciences
New York University, 100 Washington Square East, NY 10003
DNA
Translation
RNA Protein
Sequence
Transcription
FOLDING
Native State
Unfolded State
Misfolded State
Why we cannot sample conformations
This is our potential energy surface !!
“Rough” energy landscape
Levinthal paradox:
Protein with 100 amino acids has 10 50 possible conformations!
“Folding funnel”
Adiabatic free-energy dynamics (AFED)
With: Lula Rosso (Imperial College) and Jerry B. Abrams (BU)
Small subset of collective variables of interest
Distances r
Denote these coordinates as q
α
( )
α
= 1,..., n
Dihedral angles:
φ ψ
Adiabatic free-energy dynamics
L. Rosso and MET Mol. Simulat . 28 , 91 (2002); L. Rosso, P. Minary, Z. Zhu, MET J. Chem. Phys . 116 , 4389 (2002)
In a transformation to generalized coordinates: q
α
= q
α r r
N
α = N r i
= r i q q
3 N
)
Suppose first n are of particular interest.
P s s n
=
=
∫
d N p d N r e − β H
α n
∏
= 1
δ q
α r − s
α
)
∫
d N p d q e − β H p
α n
∏
= 1
δ ( q
α
− s
α
)
H =
1
2 p T M − 1 p + V ( ) H =
1
2 p T M − 1 p + V ( ( )) − kT ln ( )
Adiabatic and temperature conditions: m
1,..., n m n + 1,...,3 N
T s
T
Free-energy surface: A s s T = − kT s ln P adb
( ,..., )
Adiabatic Free-Energy Dynamics
L. Rosso and MET Mol
.
Simulat
.
28 , 91 (2002); L. Rosso, P. Minary, Z. Zhu, MET J. Chem. Phys . 116 , 4389 (2002)
Equations of motion:
α = α m q =
α
+ T m q =
α
+ T
Analysis using the Liouville operator:
S. Samuelson and G. J. Martyna, J. Chim. Phys . 94 , 1503 (1997)
D. Marx, MET, G. J. Martyna, Comput. Phys. Comm . 118 , 166 (1999)
L. Rosso, P. Minary, Z. Zhu, MET J. Chem. Phys . 116 , 4389 (2002)
L. Rosso and MET Mol. Simulat . 28 , 91 (2002)
J. B. Abrams, L. Rosso and MET J. Chem. Phys . 125 , 074115 (2006) (solvation free energy)
J. B. Abrams and MET J. Phys. Chem. B 112 , 15742 (2008)
Adiabatic Free-Energy Dynamics
Free energy surface at temperature T : e − β A q q n
β
=
∫
d 3 q e − β V q q
3 N
)
Free energy surface at temperature T s
: e − β s
A q q n
β = e − β A q q n
β
s
=
=
∫
d 3 q e − β V q q
3 N
)
P adb q q n
β β
s
)
s
Free energy profile given by:
A q q n
β
= −
kT s ln P adb q q
β β
L. Rosso, P. Minary, Z. Zhu, MET
J. Chem. Phys . 116 , 4389 (2002)
Toy model:
( , ) =
(
2 − a 2
) 2
+
1
2 ky 2 + λ xy
C ( ) = x (0) ( )
C ( ) = y (0) ( )
Conformational sampling of the solvated alanine dipeptide
[L Rosso, J. B. Abrams and MET J. Phys. Chem. B 109 , 2099 (2005)]
φ ψ
Meta 1
Time FF
5 ns CHm27
β α
R
1.0
0.0
C7 ax
4.8
US 400 ns CHm22 0.3
0.0
3.9
α
L
7.4
4.4
AFED 4.7 ns CHm22 0.2
AFED T
φ , ψ
= 5T, M
φ , ψ
= 50M
Umbrella Sampling 50 ns
C
A ( φ , ψ )[kcal/mol]
α
R
C7 ax α
L
4.7 ns
CHARMm22
0.0
4.6
8.2
1. Laio and Parrinelli, PNAS (2002)
Ensing, et al. ACR 39 , 73 (2005)
β
ψ
β
φ
α
R
N-acetyl-tryptophan-methylamide (NATMA)
• Selective IR excitation of molecules from jet gives molecules excess energy.
• Excess energy causes isomerization.
• Collisions lead to cooling.
• UV Probe collision-free cooled populations.
Dian, et al . Science 296 , 2369 (2002)
Dian, et al . JCP 117 , 10688 (2002)
Dian, et al. JCP 120 , 133 (2004)
Dian, et al .
JCP 120 , 9033 (2004)
C 5
C 7 eq
α
R
C 7 ax
Ramachandran Surfaces
J. B. Abrams and MET (in preparation)
AMBER95
T
T
Simulation Details
= 423 K
= 5 T
M = 50 M
C
2.0 fs with r-RESPA
CHARM22
FFs: CHARMM22, AMBER95, OPLS-AA
GGMT Thermostats
[Liu, et al. JCP 112 , 1685 (2000)]
C 5
C 7 eq
α
R
C 7 ax
C 5
C 7 eq
α
R
C 7 ax
OPLS-AA
Expt. C5 (0.0), C7eq (0.73 kcal/mol)
AFED without transformations/TAMD
Margliano and Vanden-Eijnden, Chem. Phys. Lett.
426 , 168 (2006)
J. B. Abrams and MET, J. Phys. Chem. B 112 , 14752 (2008)
P s s n
=
∫
d N p d N r e − β H
α n
∏
= 1
δ q
α r − s
α
)
Write δ -functions as product of Gaussians:
P s s n
=
κ →∞
βκ
2 π
n / 2
∫
d N p d N r e − β H n
exp
α
∏
= 1
−
βκ
2
( q
α
− s
α
) 2
Introduce uncoupled Gaussian integrations:
P s
1 s =
∫
N p d N r d p
− β
H +
α n
∑
= 1 p 2 s
α
2 m
α
+
κ
2
α n
∑
= 1
( q
α
− s
α
) 2
AFED without transformations/TAMD
Now impose adiabatic conditions on { s , p s
}: T s
T , m
α
>> m
Distribution generated:
P
κ
(adb) s
1 s ∝
∫
s exp
− β s
α n
∑
= 1 p 2 s
α
2 m
α
[
Z s s
] s
Z s
1 s =
∫
d N p d N r exp
− β
H +
α n
∑
= 1 p 2 s
α
2 m
α
+
κ
2
α n
∑
= 1
( q
α
− s
α
) 2
Free energy surface: A s s n
A s s n
= − kT s ln P
κ
(adb) s s n
=
κ →∞
A s
1 s
Toy model:
( , ) =
(
2 − a 2
) 2
+
1
2 ky 2 + λ xy
ς =
1
N bins
N i bins
∑
= 1
[
P x t − P exact x
]
Preliminary results for N-acetyl-(AAAAAA)-methylamide
Simulation Details
T m s
=
=
300 K, T s
= 600 K
150 m
C
κ = × i -2
(5 r-RESPA steps used for harmonic coupling)
6.0 fs
FF: AMBER95, 698 TIP3P waters
GGMT Thermostats
Run length = 5 ns
Minimum: α
R
Collective variables:
Radius of gyration of backbone:
Number of hydrogen bonds:
R
G
=
1
N b i
N b ∑
= 1
r i
−
1
N b
N j
∑ b
= 1 r j
2
N
H
=
N
O
N
∑∑
N i = 1 j = 1
1
1 −
−
(
( r r r r j j
/
/ d
0 d
0
)
)
12
6
α
R
Free-energy surface of alanine hexamer
Spatial-Warping Transformations
With: Glenn J. Martyna (IBM), Zhongwei Zhu ( Wall Street ) , Peter Minary (Stanford)
Science , December 22, 2006
x
.
Molecular dynamics in a double well
V ( x ) x p
= p m
= − dV
= dx
+ time
Crossing the barrier is a rare event!
Spatial-warping transformations
Z. Zhu, MET, S. O. Samuelson, G. J. Martyna, Phys. Rev. Lett . 88 100201 (2002)
V ( x ) Hamiltonian:
2
= +
2 p m x = − a
Canonical partition function:
Q =
∫
dp dx exp
− β
p 2
2 m
+
Change the integration variables: u
=
( )
= − +
∫
− x a dy e − β
Transformed partition function:
Q =
∫
dp du exp
− β
p 2
2 m
+
β (
− 1
( )) du
(
− 1 ( )
) (
− 1 ( )
)
Spatial-warping transformations (cont’d)
Choosing the transformation:
=
V x
0 Otherwise x = − a
V ( x )
MD: u = p
= + m
Reference potential spatial warping (REPSWA)
( ) − ( )
= − d du
(
− 1
( − 1 ( )) − ( − 1 ( ))
( )
) (
− 1 ( )
)
Difference from bias potential methods
Bias potential needs the unbiasing factor:
Q =
∫
dp dx exp
−
β
p 2
2 m
+ ( ) − ( )
exp[ −
β
( )]
Spatial-warping transformations:
Q =
∫
dp du exp
− β
p 2
2 m
+
(
− 1 ( )
) (
− 1 ( )
)
System naturally moves on the smoother surface. No unbiasing needed.
V ‡
5kT
No Transformation Transformation
10kT
10kT
How to do biomolecules and polymers
P. Minary, MET, G. J. Martyna SIAM J. Sci. Comput . 30 , 2055 (2007)
Sampling hindered by barriers in the dihedral angles:
V
Ramachandran dihedral angles (alanine dipeptide):
φ
ψ
ψ
φ
Dihedral angle transformations
Transformed angle:
φ
u
= φ
min
+
φ
∫
φ min
− β V χ
φ min
V
P. Minary, G. J. Martyna and MET SIAM J. Sci. Comp. 30 , 2055 (2008) r
6
V φ = V tors
φ S φ + α
∑
i ∈ nn
V n.b.
r φ r − r i
S r
4
φ r − r i
|)
A 400-mer alkane chain
No Transformation REPSWA Transformations
RIS Model value: 10
Replica-exchange Monte Carlo (Parallel Tempering)
Swendsen and Wang, Phys. Rev. Lett.
57 , 2607 (1986)
T
M
T
M − 1
T
1
Basic Algorithm ( Parisi, Whittington, Nemoto )
A. Create M replicas of a system with temperatures T
1
< T
2
< T
M
B. Run molecular dynamics on each replica and periodically attempt neighbor exchanges:
A ( r k + 1 , r r r k + 1 = e
−∆
10-20 Replicas
5%-20% Exchange prob.
∆
, + 1
= ( β k
− β k + 1
)
V r − V ( r k + 1) )
Comparison for a 50-mer chain using TraPPE
Martin and Siepmann, J. Phys. Chem. B 102 , 2569 (1998)
PT replicas = 10; 300 < T <1000 PT exchange prob. = 5%
Nonbonded interactions off
(dihedral angle)
Comparison for a 50-mer chain using CHARMM22
REMC/PT replicas = 10; 300 < T < 1000
Nonbonded interactions off
Comparison for 50-mer using CHARMM22 all interactions
Conclusions and Questions
1.
Spatial-warping transformations lead to substantial gains in conformational sampling over ordinary MD and REMC.
Future: Combine two approaches to gain advantages of both
Still need transformations for hydrogen bonds.
2.
Adiabatic dynamics can be used to explore and generate free-energy hypersurfaces efficiently.
3.
Use of external variables [TAMD (Maragliano and Vanden-Eijnden)/ d-AFED (Abrams and Tuckerman)] eliminates the need for explicit variable transformations.
4. Simple mass scaling leads to some enhancement of conformational sampling
Future: Combine REMC with REPSWA – advantages of both (I-Chun Lin).
How does light water help in all of these methods?