SIGNAL-GUIDED SEQUENTIAL ASSEMBLY OF NANO-BIO-COMPONENTS IN THE COMPLETELY PACKAGED MICROFLUIDIC ENVIRONMENT
advertisement
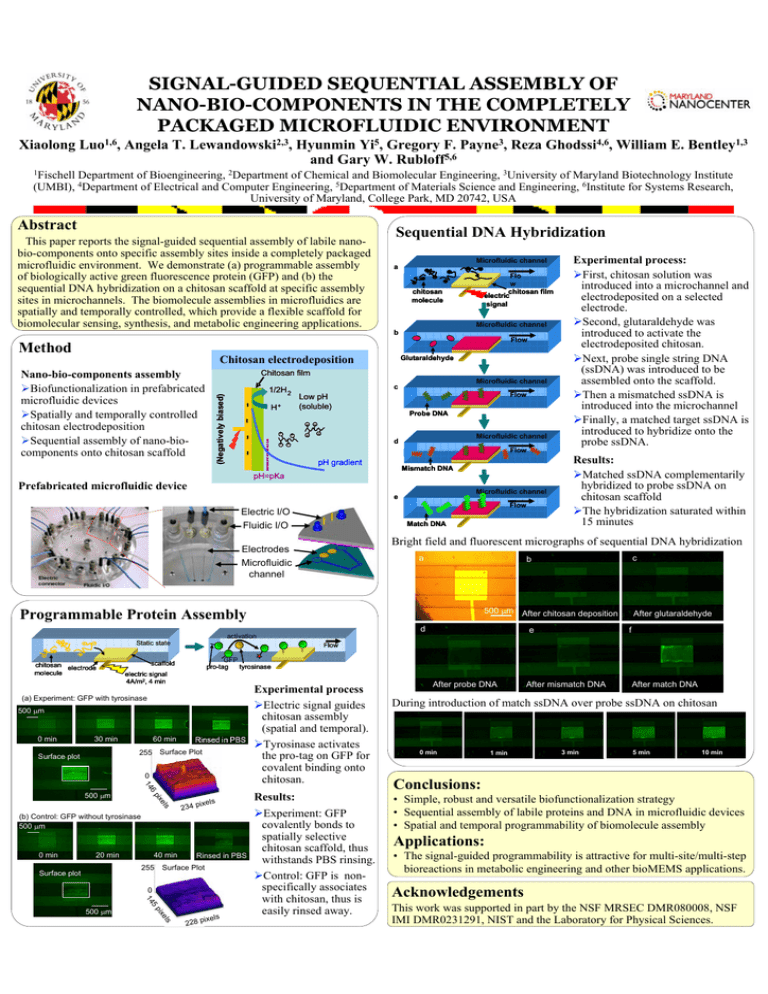
SIGNAL-GUIDED SEQUENTIAL ASSEMBLY OF NANO-BIO-COMPONENTS IN THE COMPLETELY PACKAGED MICROFLUIDIC ENVIRONMENT Xiaolong Luo1,6, Angela T. Lewandowski2,3, Hyunmin Yi5, Gregory F. Payne3, Reza Ghodssi4,6, William E. Bentley1,3 and Gary W. Rubloff5,6 1Fischell Department of Bioengineering, 2Department of Chemical and Biomolecular Engineering, 3University of Maryland Biotechnology Institute (UMBI), 4Department of Electrical and Computer Engineering, 5Department of Materials Science and Engineering, 6Institute for Systems Research, University of Maryland, College Park, MD 20742, USA Abstract This paper reports the signal-guided sequential assembly of labile nanobio-components onto specific assembly sites inside a completely packaged microfluidic environment. We demonstrate (a) programmable assembly of biologically active green fluorescence protein (GFP) and (b) the sequential DNA hybridization on a chitosan scaffold at specific assembly sites in microchannels. The biomolecule assemblies in microfluidics are spatially and temporally controlled, which provide a flexible scaffold for biomolecular sensing, synthesis, and metabolic engineering applications. Sequential DNA Hybridization Microfluidic channel a Flo w chitosan film electric signal chitosan molecule Microfluidic channel b Flow Method Chitosan electrodeposition Glutaraldehyde Chitosan film H+ Microfluidic channel c Flow Low pH (soluble) Probe DNA + + + + + Microfluidic channel d + + + (Negatively biased) 1/2H 2 - - - - Nano-bio-components assembly ¾Biofunctionalization in prefabricated microfluidic devices ¾Spatially and temporally controlled chitosan electrodeposition ¾Sequential assembly of nano-biocomponents onto chitosan scaffold Flow pH gradient Mismatch DNA pH=pKa Prefabricated microfluidic device e F F F Electric I/O Fluidic I/O Microfluidic channel F Flow Match DNA a 500 μm After chitosan deposition d e activation Static state 500 μm 60 min 255 Surface Plot 0 14 e ls pix ixels 234 p (b) Control: GFP without tyrosinase 500 μm 0 min 20 min 40 min 255 Surface plot Rinsed in PBS Surface Plot 0 5 14 pix 500 μm Experimental process ¾Electric signal guides chitosan assembly (spatial and temporal). ¾Tyrosinase activates the pro-tag on GFP for covalent binding onto chitosan. 6 500 μm f GFP tyrosinase pro-tag scaffold electric signal 4A/m2, 4 min 30 min After glutaraldehyde Flow (a) Experiment: GFP with tyrosinase Surface plot c b Programmable Protein Assembly 0 min Results: ¾Matched ssDNA complementarily hybridized to probe ssDNA on chitosan scaffold ¾The hybridization saturated within 15 minutes Bright field and fluorescent micrographs of sequential DNA hybridization Electrodes Microfluidic channel chitosan electrode molecule Experimental process: ¾First, chitosan solution was introduced into a microchannel and electrodeposited on a selected electrode. ¾Second, glutaraldehyde was introduced to activate the electrodeposited chitosan. ¾Next, probe single string DNA (ssDNA) was introduced to be assembled onto the scaffold. ¾Then a mismatched ssDNA is introduced into the microchannel ¾Finally, a matched target ssDNA is introduced to hybridize onto the probe ssDNA. els ixels 228 p Results: ¾Experiment: GFP covalently bonds to spatially selective chitosan scaffold, thus withstands PBS rinsing. ¾Control: GFP is nonspecifically associates with chitosan, thus is easily rinsed away. After probe DNA After mismatch DNA After match DNA During introduction of match ssDNA over probe ssDNA on chitosan 0 min 1 min 3 min 5 min 10 min Conclusions: • Simple, robust and versatile biofunctionalization strategy • Sequential assembly of labile proteins and DNA in microfluidic devices • Spatial and temporal programmability of biomolecule assembly Applications: • The signal-guided programmability is attractive for multi-site/multi-step bioreactions in metabolic engineering and other bioMEMS applications. Acknowledgements This work was supported in part by the NSF MRSEC DMR080008, NSF IMI DMR0231291, NIST and the Laboratory for Physical Sciences.