Multicore Programming with OpenMP Exercise Notes Getting started September 7, 2009
advertisement

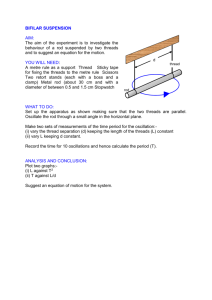
Multicore Programming with OpenMP Exercise Notes September 7, 2009 Getting started If you are using ness you can copy a tar file containing the code directly to your course account: [user@ness ˜]$ cp /home/s05/course00/multicore/openmp.tar . If you are using your own machine, then you can download the files from: http://www.epcc.ed.ac.uk/˜etg/multicore If you are using a Unix/Linux system, or Cygwin then you should download openmp.tar Unpack the tar file with the command tar xvf openmp.tar If you are using a Windows compiler then you should download openmp.zip and use standard Windows tools to unpack it. Exercise 1: Hello World This is a simple exercise to introduce you to the compilation and execution of OpenMP programs. The example code can be found in */HelloWorld/ where the * represents the language of your choice, i.e. C, or Fortran90. Compile the code, making sure you use the appropriate flag to enable OpenMP. Before running it, set the environment variable OMP NUM THREADS to a number n between 1 and 4 with the command: export OMP_NUM_THREADS=n When run, the code enters a parallel region at the !$OMP PARALLEL/#pragma omp parallel command. At this point n threads are spawned, and each thread executes the print command separately. The OMP GET THREAD NUM()/ omp get thread num() library routine returns a number (between 0 and n-1) which identifies each thread. 1 Extra Exercise Incorporate a call to omp get num threads() into the code and print its value within and outside of the parallel region. Exercise 2: Area of the Mandelbrot Set The aim of this exercise is to use the OpenMP directives learned so far and apply them to a real problem. It will demonstrate some of the issues which need to be taken into account when adapting serial code to a parallel version. The Mandelbrot Set The Mandelbrot Set is the set of complex numbers c for which the iteration z = z 2 + c does not diverge, from the initial condition z = c. To determine (approximately) whether a point c lies in the set, a finite number of iterations are performed, and if the condition |z| > 2 is satisfied then the point is considered to be outside the Mandelbrot Set. What we are interested in is calculating the area of the Mandelbrot Set. There is no known theoretical value for this, and estimates are based on a procedure similar to that used here. We will use Monte Carlo sampling to calculate a numerical solution to this problem. N.B. Complex numbers in C: Complex arithmetic is supported directly in Fortran, but in C it must be done manually. For this exercise the template has a struct complex declared. Then for a given complex number z, z.creal represents the real part and z.cimag the imaginary part. Then assigning z = z 2 + c is given by: ztemp=(z.creal*z.creal)-(z.cimag*z.cimag)+c.creal; z.cimag=z.creal*z.cimag*2+c.cimag; z.creal=ztemp; using double ztemp as temporary storage for the updated real part of z. Also, for the threshold test |z| > 2.0, use abs(z)>2.0 in Fortran, and in C use: z.creal*z.creal+z.cimag*z.cimag>4.0. The Code : Sequential The Monte Carlo method we shall use generates a number of random points in the range [(−2.0, 0), (0.5, 1.125)] of the complex plane. Then each point will be iterated using the equation above a finite number of times (say 100000). If within that number of iterations the threshold condition |z| > 2 is satisfied then that point is considered to be outside of the Mandelbrot Set. Then counting the number of random points within the Set and those outside will reveal a good approximation of the area of the Set. The template for this code can be found in */Mandelbrot/. Implement the Monte Carlo sampling, looping over all npoints points. For each point, assign z=c(i) and iterate as defined in the earlier equation, testing for the threshold condition at each iteration. If the threshold condition is not satisfied, i.e. |z| ≤ 2, then 2 repeat the iteration (up to a maximum number of iterations, say 100,000). If after the maximum number of iterations the condition is still not satisfied then add one to the total number of points inside the Set. If the threshold condition is satisfied, i.e. |z| > 2 then stop iterating and move on to the next point. Parallelisation Now parallelise the serial code using the OpenMP directives and library routines that you have learned so far. The method for doing this is as follows 1. Start a parallel region before the main Monte Carlo iteration loop, making sure that any private, shared or reduction variables within the region are correctly declared. 2. Distribute the npoints random points across the n threads available so that each thread has an equal number of the points. For this you will need to use some of the OpenMP library routines. N.B.: there is no ceiling function in Fortran77, so to calculate the number of points on each thread (nlocal) compute (npoints+n-1)/n instead of npoints/n. Then for each thread the iteration bounds are given by: ilower = nlocal * myid + 1 iupper = min(npoints, (myid+1)*nlocal) where myid stores the thread identification number. 3. Rewrite the iteration loop to take into account the new distribution of points. 4. End the parallel region after the iteration loop. Once you have written the code try it out using 1, 2, 3 and 4 threads. Check that the results are identical in each case, and compare the time taken for the calculations using the different number of threads. Extra Exercise Write out which iterations are being performed by each thread to make sure all the points are being iterated. Exercise 3: Image The aim of this exercise is to use the OpenMP worksharing directives, specifically the PARALLEL DO/parallel for directives. This code reconstructs an image from a version which has been passed through a a simple edge detection filter. The edge pixel values are constructed from the image using 3 edgei,j = imagei−1,j + imagei+1,j + image1,j−1 + imagei,j+1 − 4 imagei,j If an image pixel has the same value as its four surrounding neighbours (i.e. no edge) then the value of edgei,j will be zero. If the pixel is very different from its four neighbours (i.e. a possible edge) then edgei,j will be large in magnitude. We will always consider i and j to lie in the range 1, 2, . . . M and 1, 2, . . . N respectively. Pixels that lie outside this range (e.g. imagei,0 or imageM+1,j ) are simply considered to be set to zero. Many more sophisticated methods for edge detection exist, but this is a nice simple approach. The exercise is actually to do the reverse operation and construct the initial image given the edges. This is a slightly artificial thing to do, and is only possible given the very simple approach used to detect the edges in the first place. However, it turns out that the reverse calculation is iterative, requiring many successive operations each very similar to the edge detection calculation itself. The fact that calculating the image from the edges requires a large amount of computation, makes it a much more suitable program than edge detection itself for the purposes of timing and parallel scaling studies. As an aside, this inverse operation is also very similar to a large number of real scientific HPC calculations that solve partial differential equations using iterative algorithms such as Jacobi or Gauss-Seidel. Parallelisation The code for this exercise can be found in */Image/. The file notlac.dat contains the initial data. The code can be parallelised by adding PARALLEL DO/parallel for directives to the three routines new2old, jacobistep and residue, taking care to correctly identify shared, private and reduction variables. Hint: use default(none). Remember to add the OpenMP compiler flag. To test for correctness, compare the final residue value to that from the sequential code. Run the code on different numbers of threads. Try to answer the following questions: • Where is synchronisation taking place? • Where is communication taking place? Extra exercise Redo Exercise 2 using worksharing directives. Extra exercise (Fortran90 only) Implement the routine new2old using Fortran 90 array syntax, and parallelise with a PARALLEL WORKSHARE directive. Compare the performance of the two versions. 4 Exercise 4: Load imbalance This exercise will demonstrate the use of the SCHEDULE/schedule clause to improve performance of a parallelised code. The Goldbach Conjecture In the world of number theory, the Goldbach Conjecture states that: Every even number greater than two is the sum of two primes. In this exercise you are going to write a code to test this conjecture. The code will find the number of Goldbach pairs (that is, number of pairs of primes p1 , p2 such that p1 + p2 = i) for i = 2, . . . , 8000. The computational cost is proportional to i3/2 , so for optimal performance on multiple threads the work load must be intelligently distributed using the OpenMP SCHEDULE/schedule clause. The Code The template for the code can be found in */Goldbach/. 1. Initialise the array numpairs to zero. 2. Create a do/for loop which loops over i and sets each element of the array numpairs to the returned value of a function goldbach(2*i). This array will contain the number of Goldbach pairs for each even number up to 8000. 3. Once the number of Goldbach pairs for each even number has been calculated, test that the Goldbach conjecture holds true for these numbers, that is, that for each even number greater than two there is at least one Goldbach pair. Your output should match these results: Even number 2 802 1602 2402 3202 4002 4802 5602 6402 7202 Goldbach pairs 0 16 53 37 40 106 64 64 156 78 Table 1: Example output for Goldbach code 5 Parallelisation Parallelise the code using OpenMP directives around the do/for loop which calls the goldbach function. Validate that the code is still working when using more than one thread, and then experiment with the schedule clause to try and improve performance. Extra Exercise 1. Print out which thread is doing each iteration so that you can see directly the effect of the different scheduling clauses. 2. Try running the loop in reverse order, and using the GUIDED schedule. Exercise 5: Molecular Dynamics The aim of this exercise is to demonstrate how to use several of the OpenMP directives to parallelise a molecular dynamics code. The Code The code can be found in */md/. The code is a molecular dynamics (MD) simulation of argon atoms in a box with periodic boundary conditions. The atoms are initially arranged as a face-centred cubic (fcc) lattice and then allowed to melt. The interaction of the particles is calculated using a Lennard-Jones potential. The main loop of the program is in the file main.[f|c|f90]. Once the lattice has been generated and the forces and velocities initialised, the main loop begins. The following steps are undertaken in each iteration of this loop: 1. The particles are moved based on their velocities, and the velocities are partially updated (call to domove) 2. The forces on the particles in their new positions are calculated and the virial and potential energies accumulated (call to forces) 3. The forces are scaled, the velocity update is completed and the kinetic energy calculated (call to mkekin) 4. The average particle velocity is calculated and the temperature scaled (call to velavg) 5. The full potential and virial energies are calculated and printed out (call to prnout) Parallelisation The parallelisation of this code is a little less straightforward. There are several dependencies within the program which will require use of the ATOMIC/atomic directive as well as the REDUCTION/reduction clause. The instructions for parallelising the code are as follows: 6 1. Edit the subroutine/function forces.[f|c|f90]. Start a !$OMP PARALLEL DO / #pragma omp parallel for for the outer loop in this subroutine, identifying any private or reduction variables. Hint: There are 2 reduction variables. 2. Identify the variable within the loop which must be updated atomically and use !$OMP ATOMIC/#pragma omp atomic to ensure this is the case. Hint: You’ll need 6 atomic directives. Once this is done the code should be ready to run in parallel. Compare the output using 2, 3 and 4 threads with the serial output to check that it is working. Try adding the schedule clause with the option static,n to the DO/for directive for different values of n. Does this have any effect on performance? Exercise 6: Molecular Dynamics Part II Following on from the previous exercise, we shall update the molecular dynamics code to take advantage of orphaning and examine the performance issues of using the atomic directive. Orphaning To reduce the overhead of starting and stopping the threads, you can change the PARALLEL DO/omp parallel for directive to a DO/for directive and start the parallel region outside the main loop of the program. Edit the main.[f|c|f90] file. Start a PARALLEL/parallel region before the main loop. End the region after the end of the loop. Except for the forces routine, all the other work in this loop should be executed by one thread only. Ensure that this is the case using the SINGLE or MASTER directive. Recall that any reduction variables updated in a parallel region but outside of the DO/for directive should be updated by one thread only. As before, check your code is still working correctly. Is there any difference in performance compared with the code without orphaning? Multiple Arrays The use of the atomic directive is quite expensive. One way round the need for the directive in this molecular dynamics code is to create a new temporary array for the variable f. This array can then be used to collect the updated values of f from each thread, which can then be ”reduced” back into f. 1. Edit the main.[f|c|f90] file. Create an array ftemp of dimensions (npart, 3, 0:MAXPROC-1) for Fortran, [3*npart][MAXPROC] for C, where MAXPROC is a constant value which the number of threads used will not exceed. This is the array to be used to store the updates on f from each thread. 2. Within the parallel region, create a variable nprocs and assign to it the number of threads being used. 7 3. The variables ftemp and nprocs and the parameter MAXPROC need to be passed to the forces routine. (N.B. In C it is easier, but rather sloppy, to declare ftemp as a global variable). 4. In the forces routine define a variable my id and set it to the thread number). This variable can now be used as the index for the last dimension of the array ftemp. Initialise ftemp to zero. 5. Within the parallelised loop, remove the atomic directives and replace the references to the array f with ftemp. 6. After the loop, the array ftemp must now be reduced into the array f. Loop over the number of particles (npart) and the number of threads (nprocs) and sum the temporary array into f The code should now be ready to try again. Check its still working and see if the performance (especially the scalability) has improved. Extra exercise (Fortran only) Use a reduction clause for f instead of the temporary arrays. Compare the performance of the two versions. Exercise 8: Performance analysis N.B. these instructions are for ness only. If you wish to conduct this excercise on another machine, please speak to a demonstrator. This exercise is intended to use a profiling tool to do some performance analysis. */MolDyn4 contains a correct, but not very efficient solution to the MolDyn example. First of all, run a sequential version of the code (by compiling without OpenMP, and note the execution time. To profile the code, compile with -qp in addition to other flags. Run the code on the back end, say on 4 threads. This will generate a file gmon.out. The command gprof ./md will generate profiling information. Compute the ideal time (the sequential time divided by the number of threads) and compare it to the actual execution time. Estimate how much of the overhead is due to • Sequential code (code not in the parallel region) • Load imbalance ( mp barrierp) • Synchronisation (other routines starting with mp) Appendix: OpenMP on ness Hardware For this course you can compile and run code on the EPCC HPC Service. This is provided on a Sun X4600 system (ness) confugred as follows 8 • A 2-processor front-end used for compilation, debugging and testing of parallel jobs, and submission of jobs to the back-end. • One 16-processor and one 8-processor back-ends used for production runs of parallel jobs. Compiling using OpenMP The OpenMP compilers we use are the Portland Group compilers for Fortran 90 and C. To compile an OpenMP code, simply: Fortran (pgf90): add the flag -mp C (pgcc): add the flag -mp Using a Makefile The Makefile below is a typical example of the Makefiles used in the exercises. The option -O3 is a standard optimisation flag for speeding up execution of the code. ## Fortran compiler and options FC= pgf90 -O3 -mp ## Object files OBJ= main.o \ sub.o ## Compile execname: $(OBJ) $(FC) -o $@ $(OBJ) .f.o: $(FC) -c $< ## Clean out object files and the executable. clean: rm *.o execname Job Submission Batch processing is very important on the HPC service, because it is the only way of accessing the back-end. Interactive access is not allowed. For doing timing runs you must use the back-end. To do this, you should submit a batch job as follows, for example using 4 threads: qsub -cwd -pe omp 4 scriptfile where scriptfile is a shell script containing 9 #! /usr/bin/bash export OMP_NUM_THREADS=4 ./execname The -cwd flag ensures the output files are written to the current directory. The -pe flag requestes 4 processors. Note that with OpenMP codes the number of processors requested should be the same as the environment variable OMP NUM THREADS. This can be ensured by using: export OMP_NUM_THREADS=$NSLOTS You can monitor your jobs status with the qstat command, or the qmon utility. Jobs can be deleted with qdel (or via the qmon utility). 10