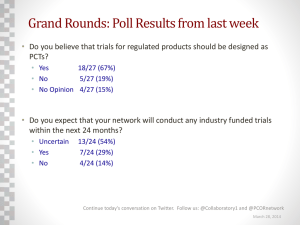
CancerGrid: developing open standards for clinical cancer informatics
advertisement

CancerGrid: developing open standards for clinical cancer informatics James Brenton1, Carlos Caldas1, Jim Davies2, Steve Harris2 and Peter Maccallum1 1 Hutchison/MRC Research Centre, University of Cambridge, Hills Road, Cambridge CB2 2XZ Oxford University Computing Laboratory, Wolfson Building, Parks Road, Oxford OX1 3QD 2 Abstract CancerGrid is a consortium developing open standards for clinical cancer informatics based on data and metadata representation, distributed service-oriented architectures, and collaborative working in virtual organisations with exceptional confidentiality constraints. In this paper we outline the unique challenges that clinical trials and translational research pose to the e-Science community, and investigate how existing technologies can best be deployed to support research with a significant clinical component. We discuss the broader picture of national and international programmes for cancer research, and evaluate the usability of the current generation of software tools in support of these programmes. 1. Introduction and background. The development of new cancer therapies is a long and hard process with low success rates and timescales measured in the decades. This might improve through molecular approaches to cancer research, but the fundamental links between the operation of cancer at the molecular level and its manifestation in the patient population will only be determined when molecular effects are observed simultaneously with the clinical effects they generate. Unfortunately clinical trials with tissue-based molecular research components are not well supported by existing information systems. The CancerGrid consortium [1] has been formed to take advantage of recent developments in Semantic Web technology, Grids, Service-Oriented Architectures (SOA), and Computer Supported Collaborative Working (CSCW). It will pursue the strategic goal of integrating clinical and research informatics infrastructures. To date CancerGrid has secured funding from the MRC for a three year technology development phase, from the EPSRC for international collaboration and from Microsoft Research to study advanced Web services applications. It is interacting with the national programme for research and clinical trials management represented by the National Cancer Research Institute (NCRI) and its international equivalents including the US National Cancer Institute’s Centre for Bioinformatics (NCICB) to ensure development will provide re-usable software toolkits and knowledge for the cancer research community. 2. New developments in cancer research practice demand a novel information architecture. Legislative changes such as the European Commission Clinical Trials Directive 2001/20/EC and the US Food and Drug Administration’s 21CFR Part 11 have put significant new pressures on clinical trials coordinators, in particular more stringent requirements on IT systems validation and on the frequency and promptness of contact with clinicians following serious adverse events. UK clinical trials units need an easy to validate set of components with inbuilt support for human to human interaction to support rapid trial deployment. Cancer researchers in the UK are committed to translational research, bridging the gap between laboratory research and clinic. However difficulties abound; individual datasets and sample collections are too small to produce statistically significant results so need to be pooled while unification can only be reliably achieved for simple measures; access to data gives rise to problems with anonymisation and re-identification of patients; and there are no standards for the collection, labelling and annotation of tissue sample sets. Translational researchers need flexible, well-structured data models and terminologies which enable data sharing while supporting the development of novel techniques. Recent successes in Cancer therapy such as Tamoxifen and Herceptin in breast cancer and Gleevec in leukaemia owe their success to an understanding of individual tumours at the molecular level; each of these compounds is only effective in individuals whose cancer cells express particular proteins. This suggests that future improvements in survival rates will be dependent on increased matching of treatments to personal genetic makeup. To fully develop this approach, larger numbers of increasingly targeted trials will be required. National cancer programmes need tools to support, monitor and direct the global knowledge base these trials will represent. 3. A prototype model-centred architecture for breast cancer trials. 3.1 CancerGrid design priniciples. Current practice in clinical trials is based on a patchwork of paper based systems and custom IT solutions for individual services. The CancerGrid project is moving its exemplar trials to an SOA-based document workflow system, with the goal of providing a Grid of generic services which can be configured to provide support for multiple trials. The past ten years have shown the great potential of custom web-delivered n-tier systems to solve individual service needs. However, this approach is expensive when each trial needs a custom solution, and to date has shown only limited success in long-term interoperability of the resulting solutions. We are taking the positive aspects of webbased delivery mechanisms, but adding the design philosophy of Model-Driven Architecture (MDA), the Semantic Web, and Grids. In the MDA approach, systems are defined in terms of their data and process models, at a level independent of the underlying implementation technology. Mappings onto specific technologies are then produced, with the help of automated tools wherever possible. In the Semantic Web approach, data models can be built from the composition and transformation of XML schemas, and formal models expressed in OWL can be used to specify components and processes. Building a clinical trial using composition of structured data elements which can be used from capture through to analysis also allows improvements both in the quality of data collected and in its eventual re-use. The Grid approach encourages systems to be built as configurable generic services, with specialised resources such as persistence mechanisms virtualised. Grid research is particularly concerned with authentication and authorisation, important in the trials area where users and services have different requirements and rights for data entry and analysis. 3.2 The prototype system. We have built a prototype system to demonstrate our approach, based on the current breast cancer therapy clinical trials tAnGo [2] and neo-tAnGo [3] for which the official documentation is already prepared and paperbased data capture is in progress. As the CancerGrid project progresses, it will be validated against the real data capture in these trials. Figure 1 shows the processes which have been implemented for tAnGo in the prototype, a subset of the set-up and data entry phases of the trial. The full system will enhance this with laboratory data capture and storage and analysis mechanisms. A generic clinical trials protocol model, a common data element repository and a vocabulary service have been created in Protégé using its XML back-end plug-in. An instance of this model, encoding the key elements of the tAnGo clinical trial protocol and its patient registration and randomisation criteria has been entered. The Protégé XML file can now be transformed using XSLT to extract an XML representation of the clinical trial protocol. This document has been further transformed to generate trial metadata, an HTML web page describing tAnGo in detail. This demonstrates how the electronic trials protocol document could be used to drive clinical trials registration and discovery systems. The description of the registration and randomisation case report form has also been transformed into two XML schemas, one the data model for data entry, and another the rules for patient eligibility in the trial. The data collection protocols accompanying the common data elements have been extracted to create HTML instructions for the completion of the case report form. Using both Adobe PDF forms and Microsoft InfoPath, the schema has been realised as a data-entry form suitable for completion by a clinician. The InfoPath example makes use of Web services to provide live access to a database of randomising hospital consultants as the form is completed. A second .NET based Web service has been created that receives XML documents from the completed forms together with XML schemas and XPATH statements derived from the protocol model. The service tests for validity, eligibility and subsequent processing requirements and Figure 1: The current CancerGrid prototype, showing the workflow driven by the trials protocol (modelled in Protégé), transformed into data and metadata for trials processes (using XSLT), which are then operated using XML-based document workflows and Web services (here using Microsoft Infopath as the user interface). demonstrates how parts of the data capture pathway for trials can be based on configurable services. The validated entry is then ready for the next stage in the trials workflow, randomisation. In the complete system the randomisation will be a separate Web service. While the XML and Web service prototype for tAnGo demonstrates the principles of document and service creation, we have in parallel provided a Zope/Plone [4, 5] based CSCW solution for the neo-tAnGo trial’s existing static Word and PDF documents. This required the development of a custom four-level Zope security model, with (1) administrators, (2) trial coordinators responsible for portal content, (3) clinical and research nurse members needing access to documents without modification privileges and (4) anonymous visitors who might include patients and the general public. In the final system many of these documents will be generated automatically from the protocol on demand, because minor changes in the protocol can still result in a significant effort by trial coordinators to update all affected content. 4. Architecture in context; the evolving landscape of clinical trials management. 4.1 Clinical vocabularies and ontologies require investment and service provision at an international level. Once the software infrastructure is in place to support trials management, the provision of high-quality curated data elements will be a determining factor in the success of the project. Collaboration with UK and international cancer organisations is required to increase the pool of available managed data resources and terminologies. CancerGrid is working with the NCICB, and plans to reuse the existing caDSR product to host standard data elements in a Web service environment, along with other components of the caBIG architecture [6]. This type of data element sharing requires a framework which imposes a common metamodel, so that a standard toolset can be used to incorporate the elements into software systems. Compliance with standards, such as ISO11179 adopted by NCICB, may be the solution. While such standards-based modelling imposes overheads, it will support UK cancer researchers’ efforts to collaborate with Australasia, the EU and North America. 4.2 Clinical trials management processes are heterogenous and evolving. The prototype system is directed at a small subset of the eventual user community, specifically trials coordinators and data entry clinical research staff. Eventually the system will need to support statisticians, bioinformaticians, regulatory authorities and commercial interests in a framework which preserves the evolving ethical and legislative constraints in the trials process. The regulatory frameworks, operating procedures and relationships between clinics, accredited trials units and national trials services are constantly evolving. The CancerGrid architecture cannot target a single fixed organisational model, but will instead build a flexible role-based model into the system and its deployment and configuration technology. 5. CancerGrid’s challenges to the current generation of Semantic Web technologies. CancerGrid will test the latest developments in MDA. Current commercial MDA tools provide good support for code skeleton generation in Java and C# but still require significant software development expertise. Moving to an XML centred system making use of XSLT reduces the need for software components – many steps in the model transformation process become metadata-defined. Adopting ontologies rather than schemas for models allows better designed systems, with aspects of formal specification built in; but the toolkits to support ontology based development are not as mature as the equivalent software design tools. Developments in this area are required; formal specification tools that non-logicians can use. The CancerGrid architecture will require a dependable persistence infrastructure. While datasets remain small, XML document repositories such as Xindice will be sufficient, but as tissue based data is included relational databases will be required. For these, an objectrelational mapping technology such as OJB could be used; but we are also considering the benefits of the OGSA-DAI approach, which would allow databases to interact with the system via XML intermediary formats. In either case we will virtualise the persistence mechanism; trials data will need to persist for up to 50 years, so it is reasonable to expect several migrations of the underlying technology during that time, while the data models should remain constant. CancerGrid will test the current limits of Web service and Grid security models. The principle of moving healthcare records over TCP/IP has been demonstrated in several previous e-Science projects such as e-Diamond [7]. However, the current WS-I Basic Profile security model encounters some issues with firewalls. Trusted communication between NHS clinics, universities and trials units will require a more advanced approach based on robust, interoperable WS-Security implementations. Authentication is also an issue. Users will need to access multiple resources outside their own institutions. Each trial could build a networked authentication system based on a technology such as LDAP, but management of that system would quickly become a significant overhead. A full solution for the UK-wide or international collaboration may require the latest developments in role-based inter-organizational security, such as the Shibboleth system [8]. CancerGrid is building systems to support human and automated workflows. We will make use of the latest developments in areas such as BPEL and Taverna/Freefluo. Bioinformatics and statistical analysis is currently done using command-line tools or custom applications. By integrating analyses as Web service workflow components, built on Grid/Web convergence technologies such as GT4, we can make toolkits available to users and also improve the specification and reproducibility of the analysis of this valuable data. 6. Conclusions Current molecular approaches to cancer research mean that we need to conduct more trials on faster timescales. The data for those trials should be collected in systems which prepare them for re-use in merged large-scale datasets. An XML-based, MDA approach has been demonstrated to adapt well to the trials data capture process. We are extending this to a Grid/SOA model of configurable services for complex virtual organizations. We plan to construct a Grid of services which can be reconfigured to match the rules and organizational structure of each clinical trial. Existing e-Science and commercial software development tools support parts of the process. As Semantic Web and Grid technologies develop, we hope be able to use them to deliver a system where a legal, ethical and organizational model can be translated directly into the deployment and configuration rules for a validated trials data management system. 7. References [1] http://www.cancergrid.org [2] http://www.cancerinformatics.org.uk [3] http://ncicb.nci.nih.gov [2] TANGO clinical trial ISRCTN 51146252 [3] Neo-tAnGo clinical trial ISRCTN 78234870 [4] http://www.zope.org [5] http://www.plone.org [6] Buetow, KH Science 308 821-824 (2005) [7] http://www.ediamond.ox.ac.uk [8] http://shibboleth.internet2.edu