How much stats does it take to time scale? Alexandre Gramfort
advertisement

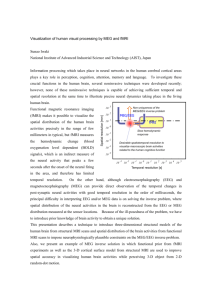
How much stats does it take to look at the brain at a millisecond time scale? Alexandre Gramfort alexandre.gramfort@telecom-paristech.fr Assistant Prof. Telecom ParisTech CEA - Neurospin, France Biomag - Aug. 2014 The overall goal invasivity Coarse spatial resolution (mm) weak EEG 20 MEG SPECT 15 nIRS 10 Challenge of the inverse pb Fine strong 5 PET fMRI sEEG MRI(a,d) 1 Fast Alexandre Gramfort 10 102 103 104 Slow Neurostats 2014 105 temporal resolution (ms) 2 EEG & MEG in a nutshell Equivalent Current Dipole V EEG recordings Equivalent Current Dipole dB/dz MEG recordings B E Neural Current (post synaptic) First EEG recordings in 1929 by H. Berger Neural Current (post synaptic) -JRVJE %FXBS IFMJVN 4FOTPST Hôpital La Timone Marseille, France Alexandre Gramfort Neurostats 2014 3 M/EEG Measurements EEG : • ≈ 32 to 100 sensors MEG : • ≈ 150 to 300 sensors Sampling between 250 and1000 Hz THM: High temporal resolution Sample EEG measurements Alexandre Gramfort Neurostats 2014 4 ains the unknown amplitudes of the sou unknown amplitudes of the sources an M/EEG Measurements: Notation e unknown amplitudes of the source ources by dx , the number of sensors by yby dx ,dthe number of sensors by d an m dm : Nb dtofby , the number of sensors d sensors x e notations, we have,dmM dt R ,dm G dm x ns, we have, M R M = dm , G dt : Nb ofRtime points dm ions, we have, M R ,G R MEG and/or EEG nge of 10, for low resolution EEG, and , for low resolution EEG, and 400, fo 10, resolution EEG, and 400 ies. for Thelow parameter d is commonly bet t 1 column parameter d is commonly between 1 t fiers used in M/EEG, the sampling rat he parameter dt is commonly betwee d dinwhen M/EEG, the sampling rate can 1 row = 1 time series recording several seconds t in M/EEG, the sampling rateofca sed 1 sensor recording several secondson of signal. 1.1 0.5 0 −0.5 en recording several seconds of sign −1.1 2D topography Alexandre Gramfort 3D topography Neurostats 2014 5 Distributed model Position 5000 candidate sources over each hemisphere (e.g. every 5mm) Space Time X sources amplitudes Scalar field defined over time Alexandre Gramfort [Dale and Sereno 93] Neurostats 2014 6 Distributed model Position 5000 candidate sources over each hemisphere (e.g. every 5mm) Space Time X sources amplitudes Scalar field defined over time Alexandre Gramfort [Dale and Sereno 93] Neurostats 2014 7 Distributed model one column = Forward field of one dipole G= EEG forward field on the electrodes GEEG GMEG MEG forward field on sensors G is the gain matrix obtained by concatenation of the forward fields Alexandre Gramfort Neurostats 2014 8 Distributed model one column = Forward field of one dipole G= EEG forward field on the electrodes GEEG GMEG MEG forward field on sensors G is the gain matrix obtained by concatenation of the forward fields Alexandre Gramfort Neurostats 2014 9 bles: M= GX+E : An ill-posed problem E ⇥ N (0, X ⇥ N (0, ve model: 2.5e−13 1.3e−13 0 E ⇥ N (0, X ⇥ N (0, E) E) X) X X) −1.3e−13 −2.5e−13 dx 10000 dm M = GX + E . 100 M = GX + E . M G are known, X is obtained by maximizing ained by maximizing the likelihood which ⇥ X = arg min ⇤M GX⇤ E Linear problem with more unknowns than the X number of equations: it’s ill-posed => Regularize rg min ⇤M GX⇤ Alexandre Gramfort E + ⇤X⇤ X Neurostats 2014 , 10 ume bles: Gaussian y= Xw+E :variables: An ill-posed E ⇥ N (0, X ⇥ N (0, dx problem Standard ⇥ N (0,notations E) statistics E) E X) Xw⇥ (or N (0,β) regression coefficients 10000 100 design matrix dm ve model: y = GX X M +E. X) M = GX + E . ained are known, byTHM: maximizing X is obtained the likelihood byM/EEG maximizing which At each time instant the inverse problem IS a regression with more variables than observations rg min ⇤M GX⇤ X = Earg + min ⇤X⇤⇤M , GX⇤ X Alexandre Gramfort ⇥ Neurostats 2014 11 E ⌅1 Inverse problem framework X = arg min ⇤M GX⇤ + ⇥⇤( ⌅ 2 2 F X X = arg min ⇤M GX⇤F , subject to ⇤(X) ⇥ Penalized (variational) formulation (with whitened data): X ⌅w,1 ⌅w,2X ⇤(X) = arg min ⇤M X Data fit 2 GX⇤F + ⇥⇤(X), ⇥ > 0 Regularization ⌅1 , ⌅2 , entropy . . . ⇥ : Trade-off between the data fit and the regularization ⇤(X) 2 T where ⇤A⇤F = tr(A A) ⌅1 , ⌅2 , entropy . . . How do you whiten data? ⌅1 ⌅2 ⌅w,1 ⌅2 Alexandre Gramfort ⌅1 Neurostats 2014 ⌅w,2 12 Baseline “noise” Data from different sensors have to be put on the same scale. 11 tt CC==T MYM Y T With whitened data the covariance would be diagonal Model selection: Log-likelihood Given my model C how likely are unseen data Y? L(Y |C) = 1 t Trace(Y Y C 2T 1 ) 1 N log((2⇡) det(C)) . 2 Higher log likelihood = better C & better whitening Cross-validation -1234.3 -1324.7 -1467.0 -1178.9 average log likelihood and select the best model We compared 5 strategies: 1. Hand-set (REG) C 0 = C + ↵I, simple, fast ↵>0 2. Ledoit-Wolf (LW) CLW = (1 µ = mean(diag(C)) ↵)C + ↵µI 3. Cross-validated shrinkage (SC) CSC = (1 ↵CV )C + ↵CV µI 4. Probabilistic PCA (PPCA) t CP P CA = HH + 2 IN 5. Factor Analysis (FA) t CF A = HH + diag( 1, . . . , D) complex, slow MEG and EEG data log likelihood scores CSC = (1 ↵CV )C + ↵CV µI or CF A = HH t + diag( 1, . . . , D) whitening based on best C Text viz 1 : whitened evoked data whitened Global Field Power ( 2 ) bles: back to M = G X + E E ⇥ N (0, X ⇥ N (0, In previous sections, the ⇥2 priors used in the penalizati fined a priori. Following the explanations in section 3.2.2 methods assume a predefined covariance matrix for the sour we will present inverse solvers that aim at designing a pri covariance matrix, i.e., the weights in the ⇥2 penalization te that the model is learned from the data [193]. E) For simplicity, we will present the following method in inverse computation. X X) The methods presented in this section use the Bayesian lem. We recall the Bayesian framework from section 3.2.2.2 sources p(X|M) = where we assume Gaussian variables: ≈10 000 dipoles data forward operator ≈100 sensors G M M = GX + E . p(M|X)p(X) . p(M) E ⇥ N (0, E) X ⇥ N (0, X) +E and an additive model: M = GX + E . If E and X noise are known, X is obtained by maximizing the X⇥ = arg min ⇤M GX⇤ E + ⇤X ained by maximizing the likelihood which X which leads to: X⇥ = rg min ⇤M X XG T (G T + E) 1 In this framework the prior is an ⇥2 norm and learning the p source covariance matrix. One may also want to learn the n that in the WMN framework, learning X consists in learn In the case where X and E are not fixed a priori, th commonly denoted M. Bayes’ rule can be rewritten: Objective: estimate M GX⇤ E + ⇤X⇤ XX given , Alexandre Gramfort XG p(X|M, M) = p(X|M,2014 M) is called the posterior. Neurostats p(M|X, M)p(X|M p(M|M) 24 Challenge: Space How do you promote sparse solutions with non-stationary sources? `21 + `1 Time Support of X Change the represensation Original STFT 50 STFT coef. [“Wavelet shrinkage” Donoho & Johnstone 94] [“Soft thresholding” Donoho 95] [Application to evoked EEG, O. Bertrand et al. 94] [Application to ST EEG, Quiroga et al. 03] etc. [Moussallam, Gramfort, Richard, Daudet, Signal Processing Letters 2014 ] Alexandre Gramfort Neurostats 2014 26 E ⇥ N (0, X ⇥ N (0, E) X) e model: G M M = GX + E . data Frequency M = GZΦ + E es: Gaussian variables: me forward operator E ⇥ N (0, X ⇥ N (0, TF coefficients E) X) Z +E Φ M = GX + E . TF coefficients TF dictionary noise Z Z inedknown, by maximizing the likelihood which are X is obtained by maximizing Objective: estimate Z given M [Gramfort et al., Time-Frequency Mixed-Norm Estimates: Sparse M/EEG imaging ⇥ with non-stationary source activations, Neuroimage 2013] E X Alexandre Gramfort Neurostats 2014 27E g min ⇤M GX⇤ + ⇤X⇤ X = arg min ⇤M , GX⇤ + Time-frequency (TF) regularization The classical approach [MNE, dSPM, sLORETA]: X̂ = arg min kM X data fit + (X), > 0 regularization 2 GXkF we propose: Ẑ = arg min kM Z GZ H 2 kF + (Z), then X̂ = Ẑ H • : is a TF dictionary (STFT) • Z : coefficients of the TF transform of the sources Advantage: localization in space, time and frequency in one step Alexandre Gramfort Neurostats 2014 28 `2 Space Space What regularization? `21 Time Time Space Space Time `1 `21 + `1 Time (Z) = (⇢kZk1 + (1 (X) = kXk21 = Alexandre Gramfort X i Neurostats 2014 s X t ⇢)kZk21 ) |xi,t |2 29 non-di↵erentiable associated to f2 . [21]: F(Z) = f1 (Z) + f2 (Z). The cost function in (2) belongs to this category. However, we need to be able to compute the proximal operator M Definition operator). Let ' : R ! R be a proper convex associated to1 f(Proximity . 2 function. The proximity operator associated to ', denoted by prox' : RM ! RM M Definition 1 (Proximity operator). Let ' : R ! R be a proper convex reads: 1 to ', 2denoted by prox : RM ! RM function. The proximity operator associated ' prox' (Z) = arg min kZ Vk2 + '(V) . reads: V2RM 2 1 2 prox (Z) = arg min kZ Vk . ' 2 + '(V) operator In the case of the composite prior in (3), the proximity is given by 2 M V2R the following lemma. In the case of the composite prior in (3), the proximity operator is given by P ⇥K Lemma 1 (Proximity operator for ` + ` ). Let Y 2 C be indexed by 21 1 the following lemma. a double index (p, k). Z = prox (⇢k.k1 +(1 ⇢)k.k21 ) (Y) 2 CP ⇥K is given for each P ⇥K Lemma 1 (Proximity operator for ` + ` ). Let Y 2 C be indexed by 21 1 coordinates (p, k) by P ⇥K a double index (p, k). Z = prox (⇢k.k10 (Y) 2 C is1given for each +(1 ⇢)k.k21 ) + coordinates (p, k) by Yp,k (1 ⇢) +@ A . Zp,k = (|Yp,k | ⇢) 01 qP 1 + 2 |Yp,k | + (|Y p,k | ⇢) ⇢) k Yp,k (1 + A . Zp,k = (|Yp,k | ⇢) @1 qP |Yp,k+| +2 0 (|Y | ⇢) p,k where for x 2 R, (x) = max(x, 0) , and by convention k 0 =0 . Proximal gradient algorithm 0derived for hierarchical + This result is a corollary of the proximity operator where for x 2 R, (x) = max(x, 0) , and by convention THM: It boils down to 2 successive thresholdings 0 =0 . group penalties recently proposed in [22]. The penalty described here can indeed This as result is a corollary of[Jenatton the proximity derived for hierarchical be seen a 2-level hierarchical structure, the resulting proximity operator etand al. operator 2011, Gramfort et al. IPMI 2011] group penalties recentlyapplying proposedthe in [22]. The penalty described here can indeed reduces to Alexandre successively ` proximity operator then the ` one. 21 1 Gramfort Neurostats 2014 30 Simulation results (part 1) 0 −20 20 25 time (ms) 30 0 −10 −20 10 35 15 (a) X ground truth Potential (mV) ECD amplitude 20 0 −20 15 20 25 time (ms) 30 Potential (mV) 20 0 −20 15 20 25 time (ms) 30 35 (g) X⇥21 40 −20 0 35 Alexandre Gramfort 50 10 50 0 −10 150 200 100 150 15 20 25 time (ms) 30 200 35 0 10 50 0 −10 −20 10 20 40 60 80 time (frame) 100 120 (f) Non-zeros of X⇥1 20 100 150 15 20 25 time (ms) 30 (h) M⇥21 = GX⇥21 20 100 time (ms) (c) M noisy (SNR=6dB) (e) M⇥1 = GX⇥21 40 −10 0 −20 10 35 0 20 (d) X⇥1 −40 10 30 (b) M noiseless 40 −40 10 20 25 time (ms) 10 ECD 15 10 ECD −40 10 20 Potential (mV) Potential (mV) 20 20 ECD amplitude ECD amplitude 40 35 200 20 40 60 80 time (frame) 100 120 (i) Non-zeros of X⇥21 Neurostats 20140 31 −40 10 Simulation results (part 2) 15 20 25 time (ms) 30 −10 Potential (mV) Potential (mV) ECD amplitude ECD amplitude 30 0 0 2020 2525 time (ms) time (ms) 3030 3535 20 25 20 25 time (ms) time (ms) 30 30 0 0 1515 2020 2525 time (ms) time (ms) 3030 3535 0100 −20200 0 2050 40 100 80 60 time (ms) time (frame) 150 100 120 200 (c)(i)MNon-zeros noisy (SNR=6dB) of X⇥21 50 50 15 15 20 25 20 25 time (ms) time (ms) 30 30 100 100 150 150 200 200 35 35 20 40 60 80 100 120 1000 time 2000 4000 (frame)3000 TF coefficient Non-zeros ⇥1 (l)(f) Non-zeros of of Z⇥X 21 +⇥1 20 Potential (mV) 120 0 0 (e) = GX⇥21 ⇥1TF (k)M M ⇥21 +⇥1 40 100 −10150 20 20 10 10 0 0 −10 −10 −20 −2010 10 (d) X⇥1 (j) X TF ⇥21 +⇥1 60 80 time (frame) 10 50 (b) M M⇥ noiseless (h) = GX⇥21 21 35 35 40 20 0 −20 −20 1010 Potential Potential(mV) (mV) 15 15 20 (f) Non-zeros of X⇥1 −10 −10 1515 200 35 1010 (a) X ground (g) X⇥21truth ECD amplitude 20 25 time (ms) 2020 −20 −20 ECD ECDamplitude amplitude 15 (e) M⇥1 = GX⇥21 2020 100 150 −20 10 35 4040 40 40 20 20 0 0 −20 −20 −40 −4010 10 ECD 0 (d) X⇥1 −40 −40 1010 50 ECD ECD −20 10 Potential (mV) ECD 0 Potential (mV) ECD amplitude 20 0 ECD 20 Simulations results with SN 10 Fig. 3. R = 6 dB. (a) simulated50 source activations. (b) 0 100 Noiseless simulated measurements.0(c) Simulated measurements corrupted by noise. (d−20 −10 150 e-f) Estimation with 1 prior. (g-h-i) Estimation with 21 prior [8]. (j-k-l) Estimation −40 −20 200 10 15 20 25 30 35 10 15 20 25 30 35 20 40 60 80 100 120 with composite TF (f-i-l) show the sparsity patterns2014 obtained by time (ms)prior. time (ms)Neurostats timethe (frame) 3 different Alexandre Gramfort 32 MEG Auditory data Protocol: 50 epochs of auditory tones in left ear (305 MEG, 59 EEG channels) (a) MEG data (Gradiometers only) (b) GX?TF-MxNE (explained data) (c) X?MxNE (d) X?TF-MxNE 16ms Alexandre Chronometry Fig. 6. Results obtained with TF-MxNE and MxNE for an auditory stimulation (left hear stimulation) Gramfort Neurostats 2014 A1c A1i 33 MEG Visual data Protocol: 50 epochs of visual flash in left hemi-field (305 MEG, 59 EEG channels) V1 V2d 7ms Alexandre Gramfort Neurostats 2014 34 LCMV dSPM MNE Software for MEG and EEG http://www.martinos.org/mne http://www.github.com/mne-tools MNE software for processing MEG and EEG data, A. Gramfort, M. Luessi, E. Larson, D. Engemann, D. Strohmeier, C. Brodbeck, L. Parkkonen, M. Hämäläinen, Neuroimage 2013 Alexandre Gramfort Neurostats 2014 36 Development of the MNE software U. Wash. Berkeley UCSF Sheffield Cambridge Harvard Paris NYU Aalto Ilmenau Juelich Gratz http://martinos.org/mne/stable/contributing.html References 1 po std oc Gramfort et al., Mixed-norm estimates for the M/EEG inverse problem using po accelerated gradient methods, Physics in Medicine and Biology, 2012 sit i Gramfort et al. Time-frequency mixed-norm estimates: Sparse M/EEG imaging with non-stationary source activations, NeuroImage, 2013 Strohmeier et al., Improved MEG/EEG source localization with reweighted mixed-norms, International Workshop on Pattern Recognition in Neuroimaging (PRNI), 2014 Engemann, D.A., Gramfort, A.. Automated model selection in covariance estimation and spatial whitening of MEG and EEG signals. (submitted) Collaborators: M. Hämäläinen, MGH / Harvard / MIT, Boston D. Strohmeier & J. Haueisen, TU Ilmenau, Germany D. A.Engemann, CEA Neurospin, ICM, Paris, France M. Kowalski, L2S, Univ. Paris-sud on