732A35 1
advertisement

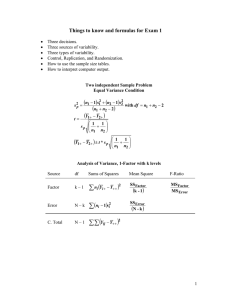
732A35 1 This method was introduced at the last lecture. Cell means model 𝑌𝑖𝑗𝑘 = 𝜇𝑖𝑗 + 𝜀𝑖𝑗𝑘 Factor effects model 𝑌𝑖𝑗𝑘 = 𝜇∙∙ + 𝛼𝑖 + 𝛽𝑗 + 𝛼𝛽 𝑖𝑗 + 𝜀𝑖𝑗𝑘 732A35 2 Method of least squares or maximum likelihood is used to minimize: 𝑄= 𝑌𝑖𝑗𝑘 − 𝜇𝑖𝑗 𝑖 𝑗 2 𝑘 or 𝑄= 𝑌𝑖𝑗𝑘 − 𝜇∙∙ − 𝛼𝑖 − 𝛽𝑗 − 𝛼𝛽 𝑖 𝑗 2 𝑖𝑗 𝑘 732A35 3 𝑆𝑆𝑇𝑂 = 𝑌𝑖𝑗𝑘 − 𝑌∙∙∙ 𝑖 𝑗 𝑘 𝑆𝑆𝑇𝑅 = 𝑛 𝑌𝑖𝑗∙ − 𝑌∙∙∙ 𝑖 𝑆𝑆𝐸 = 𝑗 𝑘 2 𝑗 𝑌𝑖𝑗𝑘 − 𝑌𝑖𝑗∙ 𝑖 2 2 2 𝑒𝑖𝑗𝑘 = 𝑖 𝑗 732A35 𝑘 4 SSTR can be partitioned: 𝑆𝑆𝐴 = 𝑛𝑏 𝑌𝑖∙∙ − 𝑌∙∙∙ 2 𝑖 𝑆𝑆𝐵 = 𝑛𝑎 𝑌∙𝑗∙ − 𝑌∙∙∙ 2 𝑗 𝑆𝑆𝐴𝐵 = 𝑛 𝑌𝑖𝑗∙ − 𝑌𝑖∙∙ − 𝑌∙𝑗∙ + 𝑌∙∙∙ 𝑖 2 𝑗 732A35 5 As usual, the sum of squares are divided by their degrees of freedom: 𝑆𝑆𝐴 𝑀𝑆𝐴 = 𝑎−1 𝑆𝑆𝐵 𝑀𝑆𝐵 = 𝑏−1 𝑆𝑆𝐴𝐵 𝑀𝑆𝐴𝐵 = 𝑎−1 𝑏−1 𝑆𝑆𝐸 𝑀𝑆𝐸 = 𝑎𝑏 𝑛 − 1 732A35 6 First of all the means should be visualized in a treatment means plot (Interaction plot in Minitab). 732A35 7 To test if interactions are important, a F-test is calculated. 𝐻0 : 𝑎𝑙𝑙 𝛼𝛽 𝑖𝑗 𝐻𝑎 : 𝑛𝑜𝑡 𝑎𝑙𝑙 𝛼𝛽 𝐹∗ =0 𝑖𝑗 =0 𝑀𝑆𝐴𝐵 = 𝑀𝑆𝐸 𝐼𝑓 𝐹 ∗ > 𝐹 1 − 𝛼; 𝑎 − 1 𝑏 − 1 , 𝑛 − 1 𝑎𝑏 732A35 → 𝑅𝑒𝑗𝑒𝑐𝑡 𝐻0 8 If the interactions aren’t important, test for factor effects. Factor A Factor B 𝐻0 : 𝑎𝑙𝑙 𝛼𝑖 = 0 𝐻0 : 𝑎𝑙𝑙 𝛽𝑗 = 0 𝐻𝑎 : 𝑛𝑜𝑡 𝑎𝑙𝑙 𝛼𝑖 = 0 𝑀𝑆𝐴 ∗ 𝐹 = 𝑀𝑆𝐸 Critical value with a-1 and n-1(ab) degrees of freedom. 𝐻𝑎 : 𝑛𝑜𝑡 𝑎𝑙𝑙 𝛽𝑗 = 0 𝑀𝑆𝐵 ∗ 𝐹 = 𝑀𝑆𝐸 Critical value with b-1 and n-1(ab) degrees of freedom. 732A35 9 The methods on the following slides are used when the interaction is considered unimportant. Point estimators are: 𝜇𝑖∙ = 𝑌𝑖∙∙∙ 𝑎𝑛𝑑 𝜇∙𝑗 = 𝑌∙𝑗∙ Unbiased estimators of the variances: 𝑀𝑆𝐸 𝑀𝑆𝐸 2 2 𝑠 𝑌𝑖∙∙ = 𝑎𝑛𝑑 𝑠 𝑌∙𝑗∙ = 𝑏𝑛 𝑎𝑛 Confidence interval is created with the t distribution: 𝑌 ± 𝑡 1 − 𝛼 2 ; 𝑛 − 1 𝑎𝑏 ∗ 𝑠 𝑌 732A35 10 Tukey is used when all pairwise comparisons are to be made: 𝐷 = 𝑌𝑖∙∙ − 𝑌𝑖 ′∙∙ 1 2𝑀𝑆𝐸 𝐷± 𝑞 1 − 𝛼; 𝑎, 𝑛 − 1 𝑎𝑏 ∗ = 𝑏𝑛 2 𝐷±𝑇∗𝑠 𝐷 𝐷 = 𝑌∙𝑗∙ − 𝑌∙𝑗 ′∙ 1 2𝑀𝑆𝐸 𝐷± 𝑞 1 − 𝛼; 𝑏, 𝑛 − 1 𝑎𝑏 ∗ 𝑎𝑛 2 732A35 11 If only a few pairwise comparisons is to be made, the Bonferroni method usually is the best. Instead of T, B should be used. 𝐵 = 𝑡 1 − 𝛼 2𝑔 ; 𝑛 − 1 𝑎𝑏 Where g is the number of comparisons. 732A35 12 Contrasts can also be obtained, with either the Scheffé procedure or Bonferroni (see p. 852). Be aware that the standard deviation and the multiplicator is not the same for the different factors. 732A35 13 When the interactions are important, only the treatment (cell) means should be analyzed. I only show Tukey at the slides. 𝑌𝑖𝑗∙ − 𝑌𝑖 ′𝑗 ′∙ 1 2𝑀𝑆𝐸 ± 𝑞 1 − 𝛼; 𝑎𝑏, 𝑛 − 1 𝑎𝑏 ∗ 𝑛 2 =𝐷±𝑇∗𝑠 𝐷 732A35 14 On the upcoming (last) lecture we will discuss: • Two-way ANOVA with only one case per treatment (ch 20) • Randomized complete block designs (ch 21) • Analysis of Covariance (ch 22) 732A35 15 Chapter 19 Start reading chapter 20, 21, 22 732A35 16