Genetic Diversity in Brassica
advertisement
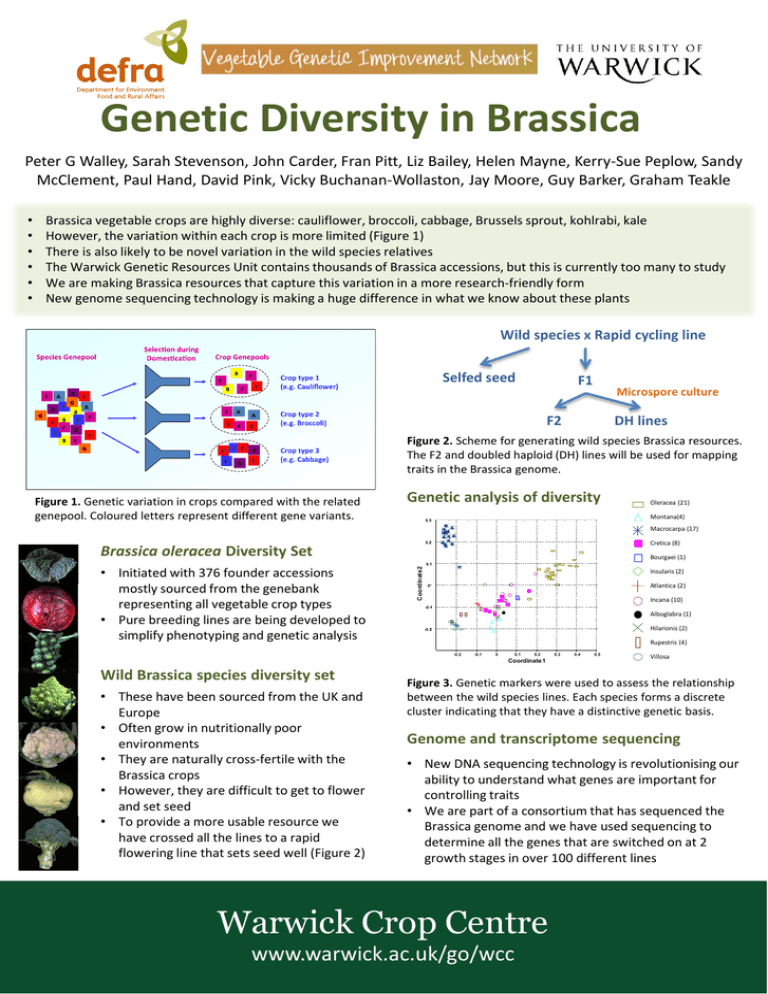
Genetic Diversity in Brassica Peter G Walley, Sarah Stevenson, John Carder, Fran Pitt, Liz Bailey, Helen Mayne, Kerry-Sue Peplow, Sandy McClement, Paul Hand, David Pink, Vicky Buchanan-Wollaston, Jay Moore, Guy Barker, Graham Teakle Brassica vegetable crops are highly diverse: cauliflower, broccoli, cabbage, Brussels sprout, kohlrabi, kale However, the variation within each crop is more limited (Figure 1) There is also likely to be novel variation in the wild species relatives The Warwick Genetic Resources Unit contains thousands of Brassica accessions, but this is currently too many to study We are making Brassica resources that capture this variation in a more research-friendly form New genome sequencing technology is making a huge difference in what we know about these plants Wild species x Rapid cycling line Selfed seed F1 Microspore culture F2 DH lines Figure 2. Scheme for generating wild species Brassica resources. The F2 and doubled haploid (DH) lines will be used for mapping traits in the Brassica genome. Figure 1. Genetic variation in crops compared with the related genepool. Coloured letters represent different gene variants. Genetic analysis of diversity Oleracea (21) Montana(4) 0.3 Macrocarpa (17) Brassica oleracea Diversity Set 0.2 • Initiated with 376 founder accessions mostly sourced from the genebank representing all vegetable crop types • Pure breeding lines are being developed to simplify phenotyping and genetic analysis 0.1 Cretica (8) Bourgaei (1) Coordinate 2 • • • • • • Insularis (2) Atlantica (2) 0 Incana (10) -0.1 Alboglabra (1) Hilarionis (2) -0.2 Rupestris (4) -0.2 -0.1 0 0.1 0.2 Coordinate 1 Wild Brassica species diversity set • These have been sourced from the UK and Europe • Often grow in nutritionally poor environments • They are naturally cross-fertile with the Brassica crops • However, they are difficult to get to flower and set seed • To provide a more usable resource we have crossed all the lines to a rapid flowering line that sets seed well (Figure 2) 0.3 0.4 0.5 Villosa Figure 3. Genetic markers were used to assess the relationship between the wild species lines. Each species forms a discrete cluster indicating that they have a distinctive genetic basis. Genome and transcriptome sequencing • New DNA sequencing technology is revolutionising our ability to understand what genes are important for controlling traits • We are part of a consortium that has sequenced the Brassica genome and we have used sequencing to determine all the genes that are switched on at 2 growth stages in over 100 different lines Warwick Crop Centre www.warwick.ac.uk/go/wcc