Lab report: Module 1 22,28,29 oct 2004. Aim:
advertisement

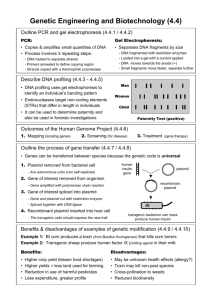
Lab report: Module 1 22,28,29 oct 2004. Aim: To extracted plasmid from bacteria (Escherichia coli) followed by transforming them again in another set of bacteria and to understand the mechanism of plasmid expression by using Polymerase Chain reaction, restriction enzymes and Agarose gel electrophoresis as tools. Plasmids: Plasmids are circular DNA, also called as extra-chromosomal DNA as they replicate independently in the bacteria. They are much smaller than bacterial DNA. Bacteria use them for exchanging useful genes. Plasmids can be made by recombinant DNA technology (cutting DNA Sequences by restriction enzymes and then sticking those using ligases enzymes [1a]) and hence they can be used as vectors. [2a] Figure1 – Diagram of a plasmid. Figure taken from http://lsvl.la.asu.edu/resources/mamajis/index.html Example of a typical plasmid is shown in the figure 1. A plasmid usually consists of about 1000 to 3000 base-pairs. Our plasmid was supposed to contain an origin of replication gene (ORI), Ampicillin resistance gene (to kill other bacteria which do not contain our plasmid) and a marker for identification example Lac Z gene. Lac Z gene when intact produces an enzyme galactosidase. This enzyme when produced catalyses X-gal (a colourless substrate present in the culture medium) into a blue precipitate and in this way Lac Z containing bacterial plasmid identified. Lac Z gene is a poly-linker, as it contains many useful restriction sites. If we insert another gene inside the Lac Z gene by using restriction enzymes then Lac Z will no longer produce galactosidase. This leads to no blue precipitate production and so only white colonies will form in place of blue. [1a] The principles and a brief outline of each step are as follows: 1. Plasmid extraction: This procedure is done by Alkali lysis method which also helps us in separating the plasmid DNA from bacterial DNA. We have two separate cultures- one containing white and the other containing blue colonies. The first step is to separate the cells from rest of the culture medium by centrifuging. We remove the supernatant and re-suspend the pellet obtained in P1 buffer (P1 contains sucrose, lysozyme and RNAse). Sucrose maintains the osmolarity and this helps prevent buffer from bursting the cells. Lysozyme helps in cutting the membrane and RNAse removes unwanted RNA. We discard the supernatant, as it may contain pieces of cell membrane. This could inhibit enzyme action on our final DNA. In the next step we add P2 (P2 contains NaOH and SDSsodium dodecyl sulphate) NaOH denatures the bacterial DNA while most of the plasmid DNA remains intact. SDS is soap and it makes holes in the cell membrane. In the next step N3 is added (contains acetic acid) which re-natures any denatured plasmid DNA from previous step. Mean while the bacterial DNA remains denatured. Then we centrifuge every thing for 20 minutes, this step ensures that denatured single strand bacterial DNA is settled at the bottom. Plasmid DNA is still in the supernatant which is transferred to silica glass bead columns. The flow is discarded and our plasmid DNA is present on the surface of the column. To the column we added PB (contains guanidine hydrochloride) which neutralizes any excess alkali. Next we added PE (ethanol with salt). Salt masks charges on DNA and thus helps ethanol to precipitate plasmid DNA. This precipitate form is later eluted with EB which contains Tris (trishydroxymethylaminomethane-buffer) and low salt. DNA was taken in another tube and stored at -20o C. 2. Transformation of Esherichia Coli: Small amount of competent cells (cells capable of accepting foreign DNA) are mixed with plasmid DNA extracted in the pervious procedure. This mixture was exposed to extreme condition of 42o C in a water bath for 2 minutes and then immediately transferring to ice. The main aim behind doing this could be that certain bacterial genes get activated which stimulates the replication as soon as favourable conditions are provided. At this stage a little bit of LB (Luria Bertani- nutrient medium nutrient medium named after the biologist) is added and whole thing was incubated for 37o C for 30 minutes which is an optimum temperature for enzymes to work. Later the cells are suspended over the surface of LB- ampicillin agar plates (Ampicillin-structurally modified penicillin derivative which interferes with bacterial cell wall synthesis and thus acts as antibiotics). Ampicillin in agar plates kills all other bacteria and except the one with our plasmid. The plates were incubated for 37o C overnight and then stored at 4o C. The plates were stored at low temperature so that bacteria can be stopped from overgrowing. 3. Polymerase chain reaction (PCR): Some cells of both blue and white type were taken from the bacteria grown in procedure 2. PCR machine was used as an incubator and each bacterial sample was heated to 100o C for 5 minutes while producing a ‘boilate’. After this step the supernatant was transferred into a fresh tube. Five clean tubes were labelled as follows: (1) sterile waterfor control. (2) diluted blue plasmid from procedure 1 (3) diluted white plasmid from procedure 1. Similarly (4) and (5) are boiled DNA from the previous step. Primer 1, Primer 2 were added along with PCR mix and template. Sterile water was later added to make up the volume. PCR mix contains Taq DNA polymerase and magnesium chloride and deoxy nucleotides e.g. (dATP, dGTP, dTTP and dCTP). Main aim was to multiply the plasmid DNA using Taq DNA polymerase (this enzyme works even at 100o C), magnesium chloride is needed to help the activity of the polymerase and the deoxy nucleotides were used by the polymerase to complete the complimentary DNA sequence. Primers are required to bind to the initial DNA base pairs and help the polymerase attachment. The PCR reaction conditions were as follows: 94o C for 5 minutes then after 30 cycles of 1 minute at 94o C (melting), 1 minute at 45o C (annealing) and 2 minute at 72o C (extension). Real time PCR is based on similar principle except that here we monitor the reaction quantitatively. So in other words we know how much is been produced at the end of the reaction. [3b] 4. Restriction Enzyme Digest: Certain enzymes are used to cut DNA at particular sequences, till now there are 200 such restriction enzymes. The enzyme which we use was called EcoRI and it use to cut DNA at GAATTC sequence and produce sticky ends e.g. CCGAATTCAA ---------- CC + GAATTCAA GGCTTAAGTT GGCTTAAG TT DNA sequence In the procedure plasmid DNA was mixed with reaction buffer, EcoRI and sterile water. The mixture was incubated for 1-2 hours at 37o C and stored at -20o C. 5. Ethanol precipitation of DNA: Plasmid DNA can also be precipitated by absolute Ethanol. Procedure include placing the mixture at -20o C overnight and then centrifuging for 30 minutes with hinge of the tube facing outwards in the rotor. Supernatant liquid was completely removed without disturbing the pallet. Glassy pallet at the side of the tube was seen. 6. Agarose Gel Electrophoresis: The principle behind this technique is that the DNA sample is place at one end of the Agarose gel and potential is applied on it such that negatively charged DNA moves towards positively charged electrode (anode). Since the Agarose is in gel form hence the separation of DNA takes place depending on the size, with smallest size DNA molecule moving furthest and largest moving slowest. This method of separation of DNA separation is called Agarose Gel Electrophoresis. TBE is use to make the Agarose into gel. TBE contains Tris and boric acid as buffers, EDTA is also added to remove divalent metal impurities like Magnesium and Calcium ions which could neutralize the charge on DNA. Ethidium Bromide is added in the gel so as to make DNA strongly fluoresces under UV illumination. DNA ladder is added along with the sample (first column shown in the photograph). It contains different size of DNA and thus helps us in getting a rough idea about the size of our products. Photograph of our gel electrophoresis: Column 1. DNA ladder, 2. PPT blue DNA, 3. PPT white DNA, 4. PPT RESTRICTION DIGEST (RD) white DNA, 5. PPT RD blue DNA, 6. blue PCR BOILED (B) DNA, 7. white PCR (B) DNA, 8. blue PCR DNA, 9. white PCR DNA. 7. DNA sequencing: The most common method for DNA sequencing is dideoxy method, also known as Sanger sequencing or chain termination method. The principle is very simple; we use dideoxy-nucleotides in place of deoxynucleotides. As shown in the figure above the former has only a hydrogen atom in place of hydroxyl group at position 3’ of the sugar. This terminates the chain during DNA synthesis by a DNA polymerase as another nucleotide cannot be added. Moreover dideoxy-nucleotides are colour tagged and since they are the last to be attached in a DNA so they keep showing the colour while moving down a gel electrophoresis. Results: DNA sequence of both Blue and White bacterial plasmid was provided after analysis. The only way we can identify the different parts of a plasmid is by recognizing the restriction enzymes sites and then searching the sites on a database e.g. blast. The blue plasmid sequence had three restriction enzyme sites so it means it has four parts. Blue Plasmid sequence> NCGNTNCTATAGGGCGAAATTGGGCCCTCTAGATGCATGCTCGAGCGGCCGCCAGTGTGA TGGATATCTGCAGAATTCGCCCTTACAACACTCGAGTCACTCAGGGGAGGCACGAACCAG GCGCACACACGTCTCTACCACCGGGCCTACCTCTACCGGCCCTAGCGACGTGACACCGGG GCGTCCCAGGTCAAGCAAGCCCCGGATGATGCGGTCGATGCGCTGCACTTCGTGGTCGAT ACGCTCCAGGAAGTCCTTCAGTTCCGGCGTGTCCGCCTTGACACGCGCCAGTGCCACGTA GCCAAGGATTCCCGCCAGCGGGTTGCCTACTTCGTGGGCGACGCCCGCGGCCAGCCGTCC GACTGTGGCCATGCGCTCCGAGAATTCTGTTGTAAGGGCGAATTCCAGCACAACTGGCGG CANTTACTAGTGGATCCGAGCTCGGTACCAAGCTTGGCGTAATCATGGTCATAGCCTTGTT TTCCTGGAAANN The white plasmid also had three sites but only two could be seen properly. In the third site a G was not recognised properly so the computer substituted it as N. With three restriction enzyme sites the white DNA plasmid can be divided into four parts. White Plasmid sequence> TTAGGGCGAATTGGGCCCTCTAGATGCCTGCTCGAGCGGCCGCCAGTGTGATGGATATCT GCAGAATTCGCCCTTCTCCAGAATTCCACCGGAAGCCCGCGCGTCCACTCAGCAACACCC AGGGCGTGCTCGAGTGGAGAAGGGCNAATTCCAGCACACTGGCGGCNNTTACTAGTGGAT CCGAGCTCGGTACCAAGNTTGGCGTAATCATGGTCATAGCTGTTTNCTGAANNN Data base search observation: All the eight sequences (four each from blue and white plasmid) where searched in the blast database individually. Fortunately only the second sequence of blue plasmid DNA could give us some good answer. It gave us three probable results shown as follows: Blast result of the second nucleotide sequence of blue plasmid DNA. Desulfovibrio vulgaris subsp Caenorhabditis elegans cosmid Streptomyces avermitilis The first result was google searched as “Desulfovibrio blue” which showed a research paper stating “A novel iron-containing blue protein, named neelaredoxin, was isolated from the sulfate-reducing bacterium Desulfovibrio gigas” [4b]. The initial conclusion we can make is that the second sequence of the blue plasmid DNA given to us produces a blue colour protein. However we can’t say form the data that the Lac Z gene (responsible for blue colour) was present or not as a plasmid is usually 1000 to 3000 base pair long and our data has only 450 base pairs roughly. Hence the data given to us is only a part of plasmid DNA. Protein search of the same sequence produced a different result from the blast database. The results showed that the DNA coded a sensor histidine kinase. At this stage it is difficult to conclude any thing for such results. Blast search of protein database using the second DNA sequence of Blue plasmid. Another surprising result was seen when we compared both blue and white sequences using blast. We found that the first and the last sequence of both were identical. Using bioedit software we compared them individually and rectified minor mistakes. The following are the rectified codes: >blue1p TAGGGCGAATTGGGCCCTCTAGATGCATGCTCGAGCGGCCGCCAGTGTGA TGGATATCTGCA >white1p TAGGGCGAATTGGGCCCTCTAGATGCATGCTCGAGCGGCCGCCAGTGTGA TGGATATCTGCA >blue4p GAATTCCAGCACACTGGCGGCAATTACTAGTGGATCCGAGCTCGGTACCA AGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGAA >white4p GAATTCCAGCACACTGGCGGCAATTACTAGTGGATCCGAGCTCGGTACCA AGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGAA After rectification we searched again in the blast database, but could not get any convincing results. Finally we joined the first and the last sequences and again searched in the blast. We found four out of five best results showing it as a part of a vector. Cloning vector pZeRO-2T Cloning vector pSG930 Cloning vector pOriR6K-zeo-ie Cloning vector pcDNA3ZEO DNA Blast results for 1 and 4 parts joined together of both blue and white plasmid. Search result from http://wishart.biology.ualberta.ca/PlasMapper/index.html for plasmid sequences. Using the rectified DNA of both first and last sequence another database was searched which was devoted only to plasmids. Search results identified HindIII 113, EcoRI, BamHI, Xbal, Pstl and many other sequences. These are usually part of poly linker present on Lac Z gene. Poly linker is also called multiple cloning site (MCS) [1a] . So we can say that the first and the last sequence may be a part of Lac Z gene. We could not find any fixed gene sequence of Lac Z gene as every company makes its own modified version of Lac Z gene. Conclusion: Firstly the sequence was not the complete plasmid (a plasmid usually contains more than 1000 base-pair). It was not clear that what part of the plasmid it was. However careful analysis showed that it had three restriction sites dividing both Blue and White sequences in four sections. Unfortunately none of them coded exactly for Lac Z gene according to the blast database search. Rectifying the data using comparison in bio edit software helped us in predicting that first and last sequences of both blue and white plasmids were same and they were part of a vector. References: Books: [1a] Lecture Notes on Molecular Medicine- John Bradley, David Johnson and David Rubenstein [2a] Molecular Biology of The Cell- Alberts [3a] Biochemistry- Donald Voet and Judith G Voet Web sites: [1b] http://lsvl.la.asu.edu/resources/mamajis/index.html [2b] http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/D/DNAsequencing.html [3b] http://www.med.sc.edu:85/pcr/realtime-home.htm [4b] http://www.ejbiochem.org/cgi/content/abstract/226/2/613 [5b] http://wishart.biology.ualberta.ca/PlasMapper/index.html