Antigenic variation in malaria involves a highly structured switching pattern Mario Recker
advertisement

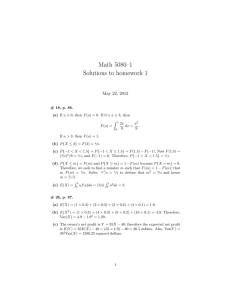
Antigenic variation in malaria involves a highly structured switching pattern Mario Recker Department of Zoology, University of Oxford Mathematical approach to (understand) malaria “the mathematical method of treatment is really nothing but the application of careful reasoning to the problems at issue.” Sir Ronald Ross 20. Aug. 1897 Ross R, 1911. The Prevention of Malaria . London: John Murray. ??? ? ? Macdonald G, 1957. The Epidemiology and Control of Malaria from McKenzie & Samba, AJTMH 2004 Most targets of protective immunity polymorphic surface proteins Development of immunity / effective vaccines hindered by extensive antigenic diversity: - mutation / recombination (genotypic change) - antigenic variation (no genotypic change) circumsporozoite protein (CSP) merozoite surface proteins (MSP) diversity variant surface antigens (VSA) www.fda.gov/CbER/blood/malaria071206sk5.gif Major multigene families: o rif o stevor o Pfmc-2TM > 150 copies per genome 30 copies per genome 13 copies per genome o var 60 copies per genome Scherf et al., Annu Rev Microbiol 2008 Sequence diversity of var genes is immense! adapted from Gardner, M. et al., 2002, Kyes, S. et al., 2002 cumulative diversity of DBLa seqnuences from Barry et al, PLoS Pathog. 2007 pairwise sharing among DBLa seqnuences Antigenic variation in P.falciparum PfEMP1 (P. falciparum Erythrocyte Membrane Protein 1) IE binding to endothelium • embedded on surface of red cell • causes severe disease through adherence to host cell receptors • important immune target IE binding to erythrocytes PfEMP1 t1 t2 Number of parasites var 1 t3 var 2 var 3 106 IE binding to dendritic cell 104 102 40 80 120 Days of infection 160 200 EM by D. Ferguson, Oxford Univ. Infected blood cells sequester in tissue capillaries EM by D. Ferguson, Oxford Univ. (Molecular) Requirement for antigenic variation - every var gene recognised as part of a family - mechanism to limit expression to a single copy - activation coinciding with silencing of previously active gene - cellular memory to avoid ‘early’ repertoire exhaustion PfEMP1 var n=1 var n = ~59 Infected RBC Result: Scherf et al., Annu Rev Microbiol 2008 succeeding waves of parasitaemia dominated by a single variant of PfEMP1 PfSir2: P.falciparum silent information regulator TPE: telomere position effect what orchestrates expression at population level? What orchestrates sequential dominance? - use mathematical models to create and test hypotheses - For example: • differences in growth rates or probabilities in switch rates (e.g. Kosinski, 1980) • differential susceptibilities assigned to variants expressing two surface proteins (e.g. Agur et al., 1989) • modifications of switch rates by ‘natural selection’ (Frank, 1999) • immunological interaction, e.g. cross-immunity (e.g. Recker et al, 2004) Increases in levels of antibodies to VSA expressed by heterologous isolates are transient and limited. 60 50 55 50 30 20 10 0 60 58 50 40 30 20 10 0 time after infection 18 Agglutinating antibody titer Percentage of infected red cells positive 40 Model assumption: each variant comprises a unique major epitope which elicits variant specific, long-lived immune response a number of minor epitopes which elicits transient, cross-reactive immune response major epitope minor epitopes Var 1 a V1 x Var 2 b V2 y Var 3 c V3 z Var 4 c V4 x Var n b Vn w The model dyi yi a zi yi a ' zi wi dt dynamics of variant i: intrinsic growth rate dynamics of specific response zi: clearance by specific response clearance by cross-reactive response dzi yi z i dt immune response proportional to antigen decay rate ’>> dynamics of transient, cross-reactive response, wi: dwi ' y j ' wi dt transient immune response proportional to antigen variants with shared epitopes Mathematical model without switching Recker et al, Nature 2004 w Vn a b V1 V2 x y b z V3 8000 3.5E+11 7000 3E+11 6000 2.5E+11 5000 2E+11 4000 1.5E+11 3000 1E+11 2000 5E+10 1000 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 contribution cross-reactive imm. response 1 peak parasitimia infection length c Model suggested that parasite-host relationship has evolved to favour some short-lived immune responses that allow the parasite to persist and the host to survive In vitro switching dynamics Horrocks et al. (PNAS, 2004) showed •on and off rates for a given variant are •on and off rates vary dramatically among Abundance dissimilar var 1 var 2 var 3 different variants time - rates appear to be intrinsic property of a particular gene - → could introduce a hierarchy of expression whereby stable variants are more prominently expressed, at least during the early phases of infection? To investigate the nature of var gene switching, generate transcription profiles for the entire repertoire in clonal parasite populations and measure the change in that profile over time stable dominance of initial variant 1st generation 2nd generation initial variant replaced change over generations determined by: X off rate t1 t2 switch bias t1 = on rate t2 off rate low low X switch bias high high - use mathematical model to determine most likely switching pathway - vit 1 1 i vit j ji vjt , i 1..n, j i off rate 1 2 3 n switch bias 0 21 31 n1 12 0 32 13 1n 23 on rate 0 0 21 31 n1 12 0 32 13 1n 1 23 2 3 use iterative approach to find ‘best-fit’ switch matrix and off-rate vector 0 n Switch matrix: variant to variant from 1 2 3 4 1 2 3 4 Switch sequence: 1→2 → 4 → 3 → Clone 3D7_AS2 Clone It_B2B even for a stable clone… Clone B12 Data provided by Dzwikowski, Frank & Deitsch Clone B10 To test the validity of this prediction, examine the var transcript distribution in Clone 2 every few generations 1 relative abundance 0.8 0.6 0.4 0.2 0 g20 g25 PFD0995c PFA0005w g30 g40 MAL7P1.56 PF10_0001 g48 g55 PFE1640w PFD0005w g60 Evolutionary conflict: protection of repertoire vs. protection against immune attack repertoire protection: immune evasion: Evolutionary conflict: protection of repertoire vs. protection against immune attack Assume var gene repertoire as a network where - nodes = variants - edges = switch / transition probabilities Task: optimise network over two traits - pathlength (= repertoire protection) - robustness (= adaptability to selection pressure) Clone 3D7_AS2 Clone It_B2B Investigate effects of hierarchical switching for in vivo dynamics naïve host highly structured switching results in (significantly?) increased length of infection. naïve host highly structured switching results in (significantly?) increased length of infection. semi-immune host sms and lattice-type pathways far more flexible in overcoming pressure from pre-existing immune responses to help set up chronic infections. Antigenic relationship between variants minor epitope 1 a b c d e minor epitope 2 u v x y z Switch sequence: (au) → (bu,av) → (cx) → (dx,cy) →… Summary • for pathogens with a limited antigenic pool, such as P. falciparum, tight control over variant expression is essential • tightly ordered gene activation requires every subsequent variant to be able to evade current immune responses and therefore may be compromised by previous infections • highly structured switching in P. falciparum has evolved as an evolutionary compromise between the protection of its limited antigenic repertoire and the flexibility to fully utilise this repertoire when needed Acknowledgements University of Oxford Department of Zoology • Sunetra Gupta • Caroline Buckee • Robert Noble THE WEATHERALL INSTITUTE OF MOLECULAR MEDICINE • Chris Newbold • Andrew Serazin • Sue Kyes • Zóe Christodoulou • Robert Pinches • Sam Kinyanjui • Pete Bull • Kevin Marsh