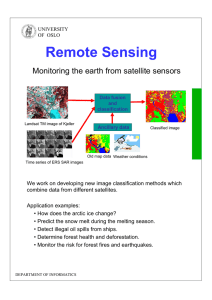
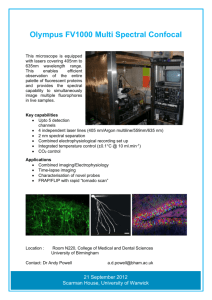
Understanding the Mesoscale Structure of the C. elegans Brain Network Dragana M.Pavlovic
advertisement

Understanding the Mesoscale Structure of the C. elegans Brain Network Prof. Edward of Warwick, Dept. of Statistics, Coventry, UK; Cambridge, UK; 3 GlaxoSmithKline, 2 University of Cambridge, Brain Mapping Unit, Dept. of Psychiatry, Differences in Community Estimation 9 4 5 8 3 4 7 5/6 4 2 3 3 2 2 1 1 1 1 2 3 4 5/67 8 9 1 2 Analysis We apply all 3 methods to the C. elegans neural network, composed of 279 non pharyngeal neurons and of 2287 undirected edges, and we use the additional functional and anatomical measures to evaluate the estimates of its community structure. For the quantitative ground truth measures, we use the Intra Class Correlation (ICC) to compare the variance explained by each community estimates. For the categorical ground truth measures, we use the Adjusted Rand Index (ARI) to compare the similarity. References [1] Daudin, Picard, Robin A mixture model for random graphs, Statistics and computing, (2008). [2] Newman, Detecting community structure in networks,The European Physical Journal B-Condensed Matter and Complex Systems, vol. 38, (2004). [3] Blondel, Guillaume, Lambiotte and Lefebvre Fast unfolding of communities in large networks, Journal of Statistical Mechanics: Theory and Experiment, vol. 10, (2008). [4] Dobson, An introduction to generalised linear models (2001). [5] Hubert and Arabie, Comparing partitions, Journal of classification, vol. 2, (1985). [6] Varshney, Chen, Paniagua, Hall, and Chklovskii, Structural properties of the Caenorhabditis elegans neuronal network, PLoS computational biology, vol. 7, (2011). 4 5 1 2 3 4 Figure 4: Spectral. Network Compression and Degree Distribution with ERMM Block 1 1.00 Block 2 18 Block 9 10 50 23 11 Block 8 Block 3 9 Dr aft 12 9 10 17 11 31 8 Block 7 0.01 28 38 Empirical Fitted 14 8 51 25 100 Block 6 46 80 30 Block 4 40 Block 5 1 Figure 5: ERMM connectivity structure. 10 Degrees 100 Figure 6: ERMM’s fitted degree distribution. Qualitative Assessment 0.6 Methods ERMM Louvain algorithm Spectral algorithm 0.4 ICC The ERMM treats the communities (blocks) and their mutual connections as mini Erdős-Rényi models, represented in the likelihood with different proportions. For a given number of communities Q, a variational approach is used to approximate the likelihood, while the Integrated Classification Likelihood (ICL) is used to compare the optimised likelihoods over different Q. The final result is an estimate of Q and the partition, visualised as a reorganised adjacency matrix. The deterministic methods like the Fast Louvain and Spectral algorithms define community as a group of highly connected nodes whose between group connections are very small. Both algorithms are devised to maximise the modularity but use different strategies to find its maximum. For example, the Fast Louvain algorithm uses a greedy approach, while the Spectral algorithm uses eigenvalues of the modularity matrix to find the optimal partition. 3 Figure 3: Louvain. Figure 2: ERMM. 31 Methods Dr. Thomas E. 1 Nichols Clinical Unit Cambridge, Addenbrooke’s Hospital, Cambridge, UK. Introduction Recently, there has been much interest in mesoscale structure of networks such as: their organisation into communities and core and periphery. However, it is often difficult to disambiguate the relationship between these two types of mesoscale structure or, indeed, to summarise the full network into the relationships between its mesoscale Figure 1: Nerve constituents. Here, we use a stochastic blockmodel aptracts C. elegans. proach Erdős-Rényi Mixture Model (ERMM)[1] for community estimation and compare this to the much more widely used deterministic methods such as: Louvain [3] and Spectral [2] algorithms. We use the Caenorhabditis elegans (C. elegans) [6] connectome (Fig. 1) as a model system in which biological knowledge about each node or neuron can be used to validate the functional relevance of the communities obtained. 2,3 T.Bullmore , 1−CDF 1 University Dr. Petra 2 E.Vertes , 0.2 0.3 Methods 0.2 ERMM ARI Dragana 1 M.Pavlovic , Louvain algorithm Spectral algorithm 0.1 0.0 0.0 ALL ALS AD BT BTD LD FC Figure 7: ICC scores for the Anatomical location (longitudinal) (ALL), Anatomical location (sectional) (ALS), Anatomical distance (AD), Birth time (BT), Birth time difference (BTD) and Lineage distance (LD). GC Figure 8: ARI scores for Functional Classification (FC) and Ganglion Classification (GC). Results The optimal ERMM fit consists of 9 classes, while the fits of Louvain and Spectral algorithms consist of 5 and 4 communities, shown in Fig. 2-4 as the reorganised adjacency matrices. The ERMM finds dense blocks on the diagonal, but but also a range of off-diagonal patterns. Note how blocks 5&6, with tight inter-connections and numerous external connections, form a core-periphery structure. Surprisingly, even though blocks 5&6 fit the standard notion of “community" they are not identified by the determinist algorithms. Furthermore, ERMM fit provides a compressed view of the C. elegans network (see Fig.5) and a faithful approximation of the degree distribution (Fig. 6). To score the quality of each fit, we show the ICC scores (Fig. 7) across the known biological features characterising nodes and edges. Here, we see that the ERMM fit scores consistently higher than Spectral and Louvain algorithms. In Fig. 8, however, ARI is rather low in general, with the Spectral algorithm showing slightly better similarity with the functional classifications and all methods having similar ARI for ganglion classification. Conclusion We showed that the Erdős- Rényi Mixture Model not only produces more biologically plausible communities but also that it provides an integrated picture of the full mesoscale structure (including core-periphery) and that it allows for compression of the network into a set of super-nodes and their connectivities. We expect these methods to prove useful for the analysis of other types of networks such as human brain functional connectivity.