The ERATO Systems Biology Workbench
The ERATO Systems Biology Workbench
Hamid Bolouri
ERATO Kitano Systems Biology Project
California Institute of Technology &
University of Hertfordshire, UK
Project PIs: Hiroaki Kitano and John Doyle
Software development team:
Andrew Finney, Michael Hucka, Herbert Sauro
Collaborators:
Adam Arkin (BioSpice), Dennis Bray (StochSim), Igor Goryanin (DBsolve),
Les Loew (VirtualCell), Pedro Mendes (Gepasi), Masaru Tomita (Ecell)
Acknowledgements: Mark Borisuk, Eric Mjolsness, Tau-Mu Yi
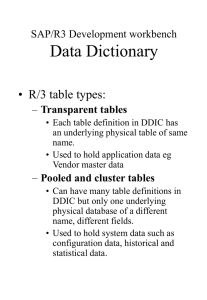
Resource Sharing, Motivation
Multistate reactions/stochastic
Reaction/Diffusion
Optimization
Bifurcation analysis
Visualization of networks
Handle large systems
Our goal: provide software infrastructure to enable sharing of simulation software (current and future) and collaboration between developers (and modelers!)
Example Workbench application: bacterial chemotaxis
Ligand binding
Signal transduction
Motion
Motor
ATP
MCPs
W
A
-ATT
+CH
3
R
+ATT
P
B
-CH
3
ATP
MCPs
W
A
P
ADP
P i
B flagellar motor
P
Y
CW
Y
Z
P i
Marlovits, Tyson, Novak, & Tyson, Biophysical Chemistry 72(1998) 169-184 k6 k3 k2 k1 cyclin cdc2 phosphorylation site
A-V model v10, April 4 th , 2000 endomes
endomes frizzled mic
mic
Maternal activator
Wnt8
Micromer e id. factor mac / veg2
mic
A1 a1
& &
& OR
&
Endo-
Mes-
TXF
& OR
GSK-3 LiCl g
& c b
MVL b
AS
Late signal from veg2 mes
mes a2 n b veg1
veg2 nN n1 mN late signal from
~ 7 th cleavage micromeres
Endoonly-
TXF
X
Zygotic
Apical N
?
N-dependent-TXF
Mes-
TXF
& &
& t
2
OR
OTX
Endo16
&
Serrate
Endo-mes
Genes
Endospecific
Genes
Mes
Genes
View from the genome, c b
=cytoplasmic b
, n b
=nuclear b
, mN=maternal N, nN=nuclear N, MVL b
AS=maternal vegetally localised b catenin activating system
ERATO Systems Biology Workbench: driving principles
• Integrate, don’t reinvent!
– integrate existing simulators
– use standard application integration methods
• object oriented, XML, Java and related technologies
• Accommodate future tools
– minimize need for ad hoc solutions
• object oriented, XML, Java and related technologies
– XML & API standards for future contributors
• Make sure contributors benefit
– symmetric plug-in infrastructure
– open source code infrastructure software
– widen user-base, but protect IPR of contributors
Systems Biology Markup Language [SBML]
• A common XML format for biochemical networks
• Enables exchange of models between simulators
• Developed in collaboration with
BioSpice, DBsolve, Gepasi, Jarnac, Ecell, StochSim, VirtualCell
• Available for public review since Sept 2000 at ftp://ftp.cds.caltech.edu/pub/caltech-erato/sbml/sbml.pdf
• Proposed extensions due 2 nd Quarter 2001
Example workbench plug-in modules
• Data filtering and preparation
– e.g. image processing, regression, clustering
• Database support
– e.g. web searching, storage management, translators, conflict resolution
• Model description tools
– scripts, languages, schematic tools
• Model preprocessing
– e.g. conserved quantities, redundancy removal
• Maths language / maths description support
• Equation solvers
– e.g. ODE, DAE, PDE, stochastic
• Analysis tools
– e.g. 2/3/4D graphing, bifurcation, MCA
• Optimization and parameter searching
Example potential plug-ins from DBsolve
• Data filtering and preparation
– regression to implicit and explicit algebraic equations
• Database support
– direct data import from WITT, MPW, KEGG
• Model description tools
– stoichiometric matrix
• Model preprocessing
– conserved quantities & redundancy removal
• Maths language / maths description support
– maths editor
• Equation solvers
– mixed ODE + NAE, LSODE
• Analysis tools
– 2D graphing, bifurcation, continuation, all steady states
• Optimization and parameter searching
– Hooke & Jeeves, Levenburg-Marquardt
Systems Biology Workbench - APIs
• APIs provided by the Workbench for simulators
– Will provide access to a spectrum of current tools
– Integration into 3 rd party simulators will require:
– SBML output
– One menu item associated with one external library call
– Available Q1 2001
– Lower level APIs for optimization, bifurcation, time-based simulation and data display will follow, Q2 2001
• APIs provided by simulators to plug into Workbench
– Existing collaborators
• no API conformance, we will interface to given APIs
• The minimum requirement:
– Either parse SBML, parse equivalent documented format or provide a model construction API
– Output some documented numeric format or structure
– Future contributors to SBW
• Standard API for independent development available Q2 2001
Systems Biology Workbench - APIs
• APIs provided by the Workbench for simulators
– Will provide access to a spectrum of current tools
– Integration into 3 rd party simulators will require:
– SBML output
– One menu item associated with one external library call
– Available Q1 2001
– Lower level APIs for optimization, bifurcation, time-based simulation and data display will follow, Q2 2001
• APIs provided by simulators to plug into Workbench
– Existing collaborators
• no API conformance, we will interface to given APIs
• The minimum requirement:
– Either parse SBML, parse equivalent documented format or provide a model construction API
– Output some documented numeric format or structure
– Future contributors to SBW
• Standard API for independent development available Q2 2001
Workbench Development Plan
Task
Design
GUI and Simulation
Engine (Sauro)
Stochastic Simulation
(Gibson)
Bifurcation analysis
(Goryanin)
Param. optimization
(Mendes)
Multi-representation simulation
Linux Port
QA, Install etc
Nov Dec Jan Feb Mar Apr May