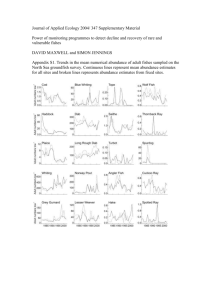
A TEST FOR COMMUNITY CHANGE USING A NULL MODEL APPROACH J S ,
advertisement

Ecological Applications, 15(5), 2005, pp. 1761–1771 䉷 2005 by the Ecological Society of America A TEST FOR COMMUNITY CHANGE USING A NULL MODEL APPROACH JACOB SCHAEFER,1,4 KEITH GIDO,2 AND MELINDA SMITH3,5 1 Department of Biology, Southern Illinois University Edwardsville, Edwardsville, Illinois 62026 USA 2Division of Biology, Kansas State University, Ackert Hall, Manhattan, Kansas 66506 USA 3National Center for Ecological Analysis and Synthesis, 735 State St., Suite 300, Santa Barbara, California 93101 USA Abstract. Quantifying patterns of temporal or spatial change in community structure is critical for assessing the impact of disturbances on biological systems and the stability of ecosystems. Detecting change in communities can be problematic, however, because of the inherent variability of systems and limitations of commonly used methods, such as similarity indices and ordination that do not explicitly test a hypothesis. Here we present empirical data to show a strong relationship between species mean abundance and variability across three broad taxonomic groups (plants, zooplankton, and fish). These statistical relationships were then used to construct null models of expected community variability that were used to test against the observed temporal change of these communities. We evaluated the ability of this approach to detect significant temporal change above that associated with random variation with nine communities (three Midwestern stream fish, three north temperate zooplankton, and three tallgrass prairie plant), each having long-term data sets and different expected levels of change. Nonrandom change was detected in 21.3% of samples from the expected low-change communities, 52.6% in moderately changed communities, and 60.4% in high-change communities. Thus, this approach was effective in detecting change over time in those communities expected to change most. By using empirical relationships between species abundance and variability, this null model approach provides ecologists and resource managers an objective tool, which can be used along with existing community indices and statistical techniques to assess the type and magnitude of community change with limited data sets. Key words: community dynamics; disturbance; null model; population variability; simulation; statistical method. INTRODUCTION Inherent variability in ecological communities can limit our ability to link community change over space or time to natural or anthropogenic disturbances (Tilman 1989, Grossman et al. 1990, Clarke and Warwick 1994, Matthews 1998, Collins 2000). In order for ecologists to relate such disturbances to community variability there needs to be more effective means for assaying the significance of the observed levels of change above that of natural or random variation. Thus, key problems facing ecologists are estimation of the amount of background variation in community structure and determination of the biological significance of deviations in community structure among samples (Grossman et al. 1990, Clarke and Warwick 1994, Philippi et al. 1998, Micheli et al. 1999, Smith 2002, Bowman et al. 2003). Long-term data at a reference site or at the same location taken prior to the disturbance are Manuscript received 28 September 2004; revised 23 December 2004; accepted 25 January 2005; final version received 9 March 2005. Corresponding Editor: T. E. Essington. 4 Present address: Department of Biological Sciences, University of Southern Mississippi, Hattiesburg, Mississippi 39401 USA. E-mail: Jake.Schaefer@usm.edu 5 Present address: Department of Ecology and Evolutionary Biology, Yale University, New Haven, Connecticut 06520 USA. ideal for addressing these problems. However, in most cases, these data are either not available or lack adequate spatial or temporal replication (Tilman 1989, Philippi et al. 1998, Collins 2000). Moreover, when longterm data are available, most techniques are not optimal for testing the null hypothesis that a community has not changed over time. In this paper, we present a null model approach to assess community change and then test the approach with both simulated (random-walk) data and empirical data from three different long-term data sets (stream fish, zooplankton, and tallgrass prairie plants). There are a number of approaches that can be used to quantify community change, including similarity indices and ordination (Washington 1984, Boyle et al. 1990, Warwick and Clarke 1991, Clarke 1993, Clarke and Warwick 1994). Each of these approaches has strengths and weaknesses depending on the questions being asked, the temporal scope of the data, and the fundamental properties of the community being examined. Consequently, researchers often rely on personal experience or some knowledge of the individual indices for selection (Jongman et al. 1995). For example, if the data set being studied consists of a small number (less than five) of samples in time, ecologists often use an index of similarity or dissimilarity to quan- 1761 1762 Ecological Applications Vol. 15, No. 5 JACOB SCHAEFER ET AL. tify community change. Quantitative similarity indices provide an intuitive measure of the differences among samples, but the behavior and sensitivity of the indices are highly dependent on the properties (e.g., abundance of dominant species) of the community (Faith et al. 1987, Boyle et al. 1990). Some authors recommend a ‘‘cut-off’’ based on mean values from a large number of comparisons to indicate a biologically-relevant change. For example, Matthews (1998) suggested that a percent similarity index (PSI) of ⬎0.6 among interannual samples in a midwestern stream fish community indicates a stable community. Because the responses of these indices change among community types, comparing values among various communities (either across geographic regions or taxonomic group) is troublesome at best. If samples are replicated and independent, standard parametric statistics can be applied to similarity indices. A variety of bootstrap or randomization techniques have also been developed to generate confidence intervals for similarity indices (Rice and Beland 1982, Ebeling et al. 1990, Solow 1993, Smith 2002) or to permute a similarity/distance matrix (Smith et al. 1990) to obtain a probability for the observed value. In addition, a multisample nonparametric correlation (Kendall’s W) can be used to test for community stability through time (Grossman 1982; but see Ebeling et al. 1990). However, a common requirement for all of these approaches is a long-term data set. In general, similarity indices as they are currently applied are of limited use unless there is a means to explicitly test the null hypothesis that a community has not changed relative to random variability in community structure. If a data set with more samples in time than species in the community is available, ordination or cluster analyses can be helpful for detecting community change. Ordination techniques allow one to quantify the amount of change as well as the directionality (direction in ordination space) of change (Clarke and Green 1988, Clarke and Warwick 1994, Legendre and Legendre 1998). Matthews (1998) used the displacement or movement of points over time in ordination space to quantify the amount and directionality of community change. More recently, Collins (2000) showed divergence in plant communities in multivariate space using autocorrelation (time-lag) analyses. However, the biggest limitation of ordination techniques is they require a large number of samples across time. Because of the problems mentioned above and the persistent need for ecologists to detect community change, we have developed a null model (Manley 1991, Gotelli 2001, Gotelli and Entsminger 2003) approach to assess community change that can be used with only a few samples. Our approach is fundamentally different from other randomization techniques in that it is based on an empirical relationship between species abundance and coefficient of variation (CV) of abundance over time for members of a community. Once quan- tified, this relationship forms the basis of a null model that predicts future community structure in the absence of any change. We therefore define community change as statistically significant change from a reference sample due to biotic or abiotic factor. Thus, our definition of change includes, among other factors, covariation among species, disturbance, and population trajectories. Our null model is based on knowledge of initial community structure (e.g., one to six reference samples) and the predicted variability of each species derived from the relationship between abundance and CV of abundance over time. Using randomly generated communities from this null model we can build frequency distributions of community change metrics (similarity indices and distances in ordination space). These distributions are then used to test the hypothesis that the community has not undergone significant change over time. Here, we present tests of this technique using random-walk simulations and long-term (14–18 yr) annual sampling records for midwestern stream fishes, a north temperate zooplankton community, and tallgrass prairie plant communities. METHODS Raw data We compiled long-term data (hereafter referred to as training samples) for three different community types to establish relationships between abundance and variation in abundance: midwestern stream fishes, north temperate lakes zooplankton, and tallgrass prairie plants. Stream fish data were taken from surveys in the Red and Arkansas River basins by the Oklahoma Department of Environmental Quality (ODEQ). The ODEQ collections included 31 sites sampled between 1977 and 1995 using a standardized sampling effort (approximately 1 h; see Pigg et al. 1998 for sample methodology). Twenty, 10-m seine hauls were taken at each site with a 3 ⫻ 1.5 m (4.7-mm mesh) heavy-leaded seine. Each site was sampled two to three times a year and all data were pooled over the year and expressed as the mean number of each species collected at each site in a year. Zooplankton data from seven Wisconsin lakes collected from 1986 to 2000 were compiled from the North Temperate Lakes Long-Term Ecological Research (NTL-LTER) database: Lake Allequash, Crystal Bog, Crystal Lake, Sparkling Lake, Big Muskellunge Lake, Trout Bog, and Trout Lake. Each lake was sampled at least three times per year at nine depths using a 2 m long Schindler-Patalas trap and with vertical tows using a Wisconsin net. As with the stream fish samples, data from all samples within a year were combined to give a mean abundance for each species in each lake during each year. At sites where yearly sampling effort changed (for example, Big Muskellunge Lake had 18 samples per year in 1986–1989 and five samples per October 2005 ASSESSMENT OF COMMUNITY CHANGE year in 1990–2000) we randomly selected a subset of samples, attempting to keep the number of samples per season equal across years. Unresolved taxa were not used in this analysis and typically consisted of a small number of rare species (usually fewer than five). Tallgrass prairie plant community data were compiled from the Konza Prairie LTER (KP-LTER) site near Manhattan, Kansas. Before 1972, sites in this area were burned at 2–3-yr intervals and grazed by cattle. Beginning in 1972, watersheds on Konza Prairie have been experimentally burned at 1-, 4-, or 20-yr intervals (Knapp et al. 1998, Collins 2000). We gathered data from two ungrazed watersheds subjected to each of these three burn regimes. Permanent sampling plots were established in 1983 and since then have been sampled in the spring and fall of each year. Species abundances were measured as percent cover as described in Gibson and Hurlbert (1987) and Collins (2000). Data were combined for spring and fall samples to give a maximum percentage cover for each species during the growing season. Relating abundance and variation in abundance The establishment of the relationship between species mean abundance and variation in abundance (from the training samples) through time is fundamental to our approach. This relationship allows us to define the predicted variability over time for each species based on its abundance in a reference sample or samples. McArdle et al. (1990) outlined the potential problems of relating species mean abundance and variability in abundance. To summarize, many measures of population variability are problematic due to the complexities of spatial and temporal scales of sampling and the dependency on mean population size. A common measure of population variability is SD(log[N]); however this measure is undefined if zero abundance data are present, something quite common in ecological data sets (Williamson 1984, McArdle et al. 1990). To correct for zero data, SD(log[N ⫹ 1]) is often used, but this measure underestimates the true variance and the more zeros in the data set the greater the bias (Anscombe 1948, McArdle et al. 1990). Coefficient of variation (CV,[SD(N)]/Nmean) is a widely used and scale-free measure of variability that does not have the same problems with zeros in the data. By plotting the CV of abundance against log(mean abundance), we can examine the relationship between a species’ (or taxon) mean abundance and variation through time. If the slope of this line is zero (a horizontal line), then variability in abundance is independent of mean abundance (McArdle et al. 1990). Because the denominator of CV is mean abundance, the relationship between log(CV) and log(mean abundance) is analogous to a residual analysis that evaluates variability as a function of an independent variable. An inverse relationship between mean abundance and CV in abundance has been demonstrated in a number of 1763 taxa including fish (Grossman et al. 1990, Winemiller 1996, Matthews 1998), mites (Ford 1938), spiders (Schoener and Spiller 1992), moths (Spitzer and Leps 1988), and mammals (Windberg 1997). Because of the generality of this relationship, we suggest these relationships can be used to predict species variability, and when used in concert with Monte Carlo simulations, can be used to evaluate the probability of community change among sample points. For each of the three community types, we calculated mean abundance and CV in abundance for all species in each of the training samples (raw data were linearly transformed [⫻10 for fish and NTL-LTER, ⫻100 for KP-LTER] so that all individual yearly species abundances were ⱖ1.0). For example, the NTL-LTER data set consisted of seven lakes (Allequash Lake, Crystal Bog, Crystal Lake, Muskellunge Lake, Sparkling Lake, Trout Bog, and Trout Lake) that had 72, 93, 80, 86, 60, and 95 total species, respectively. Therefore, there was a total of 561 species means and CVs for the NTLLTER data set from which we examined the relationship between abundance and variation in abundance. The KP-LTER and stream fish data sets consisted of 878 and 761 species–site observations, respectively. For each of the three community types, we fitted the mean log(x ⫹ 1) abundance vs. CV with an exponential decay function. There are a number of other techniques for estimating population variances (e.g., McArdle et al. 1990), but we chose this particular method because it worked well for all three of our communities. Exponential decay functions had an r2 of 0.56 for the stream fish data, 0.64 for the NTL-LTER data, and 0.65 for the KP-LTER data (Fig. 1a–c). These relationships were used to predict CV in abundance over time for a species based on its abundance in a reference sample. Testing for community change Within each of the three community types (fishes, zooplankton, prairie plants), we selected one community with a high degree of change (HC), one with moderate change (MC), and one with low change (LC) based on experimental manipulations (KP-LTER), published works, or observations by investigators (J. F. Schaefer, K. B. Gido, and J. Pigg for stream fish, J. Rusak for NTL-LTER) familiar with the communities during the respective sample periods. For stream fishes, we selected the Canadian River (HC), the Salt Fork of the Arkansas (MC), and the Salt Fork of the Red (LC); for NTL-LTER, we selected Crystal Lake (HC), Allequash Lake (MC), and Big Muskellunge Lake (LC); for the KP-LTER, we selected plots that were burned at 20-yr (HC), 4-yr (MC), and 1-yr (LC) intervals. We expected the least change in the 1-yr burn based on results presented by Collins (2000), who found these communities exhibited the lowest amount of temporal variation and turnover of species. For each of these nine communities, we used the following model to randomly generate potential com- JACOB SCHAEFER ET AL. 1764 FIG. 1. The relationship between coefficient of variation (CV) and mean abundance for each of the three communities. Solid lines represent best-fit regression lines (r2 values shown); dashed lines represent 0.5 ⫻ CV and 2.0 ⫻ CV. munities based on species abundances in a reference sample (first sample in time at one community) and their predicted CV in abundance from training samples for a community (Fig. 1). For a single randomly generated community, the model can be summarized as follows: Ai ⫽ R (x¯ i , SDi ) where Ai ⫽ the estimated abundance of species i in the randomly generated community, R is a random number drawn from a negative binomial distribution with a mean abundance of species i in the reference sample x̄i and a standard deviation (SDi) calculated from CVi. Species with an abundance of zero in a reference sample but present in later samples were included in the analysis and assigned the maximum CV for the community type and SD was calculated based on an arbitrary mean value of 1.0 (minimum abundance in transformed data). Ecological Applications Vol. 15, No. 5 Once the predicted abundance and CV for each species in a community was established, 5000 communities were generated by the model and different community change metrics were calculated using each randomly generated community and the reference sample. Sensitivity of the analysis to different levels of CV was assessed by generating an additional 5000 communities using half (0.5 ⫻ CV), and twice the expected CV (2.0 ⫻ CV). Therefore, for each community we generated a total of 15 000 randomly generated communities (Fig. 2). For all three community types, 93% (2043 out of 2200) of the CV values in training samples were within the range of 0.5(predicted CV) and 2.0(predicted CV) values (Fig. 1). Community change over time was assessed by comparing expected community structure (randomly generated communities) to target samples (all samples from a community taken after a reference sample). For each simulation, the randomly generated communities were compared to the reference sample using qualitative (Jaccard’s index) and quantitative (PSI, Bray-Curtis, and Morisita’s indices) similarity indices (Magurran 1988). Frequency distributions of each metric were compiled for each community and level of variability (CV, 0.5 ⫻ CV, and 2.0 ⫻ CV) and used to test the significance of observed similarity index values (comparing the reference sample to target samples at a community; Fig. 2). Statistically significant community change (i.e., target samples significantly different from the reference sample) occurred if the observed change for a given index was more extreme than 95% of the values of that index for the randomly generated communities (Table 1). Community change also was quantified using Euclidian distance in non-metric multidimensional scaling (NMDS) space. For each community and predicted level of CV, a NMDS ordination was performed on a matrix consisting of all target and reference samples and the 1000 (5000 was not practical due to the computational intensity of NMDS) randomly generated communities. We then calculated Euclidean distances in NMDS space between the reference sample and all randomly generated communities as well as the reference sample and the target samples. The distances between the reference sample and randomly generated communities formed a null distribution that we used to test for significant change in the target samples. Any target sample that had a Euclidean distance from a reference sample greater than 95% of the randomly generated communities was judged to have significantly changed (Fig. 2). Analyses were also performed with a second ordination technique (DCA), however results were similar to NMDS and are therefore not presented below. Accuracy of mean abundance in reference samples Because the variance estimate for each species is derived from its abundance in the reference sample or October 2005 ASSESSMENT OF COMMUNITY CHANGE 1765 FIG. 2. Flow diagram illustrating the steps involved in assessing community change. The steps shown here would assess community change from one reference sample using two metrics: (A) PSI and (B) distance in NMDS space. Results from PSI and NMDS are from a simulation of Big Muskellunge Lake using the first year as the reference sample and 1.0 ⫻ CV. The vertical line on each frequency histogram represents a 5% cutoff point used to determine significance of observed values: PSI ⬍ 0.56 or NMDS distance ⬎1.39 for a target sample would indicate significant change from the reference sample. On the NMDS plot (B), large circles represent target samples, and the black circle represents the reference sample. samples, the accuracy of our approach relies on the accuracy of this estimate. Although abundance measures for species from a single sample will reflect gross differences in abundance across species, an average abundance across multiple samples might better reflect the abundance of a species in that particular environment. Thus, to assess the accuracy of the estimated species abundance from a single sample or multiple TABLE 1. Mean similarity of randomly generated communities compared to reference samples (first-year sample) using four common similarity indices. Community type Stream Fish Zooplankton Tallgrass Prairie Community name Salt Fork of the Red River Salt Fork of Arkansas Canadian River Big Muskellunge Lake Allequash Lake Crystal Lake Annually burned 4-yr burn Unburned PSI 0.73 0.74 0.79 0.73 0.81 0.75 0.86 0.84 0.83 (0.34) (0.41) (0.42) (0.64)† (0.74) (0.55) (0.77) (0.76) (0.76) Bray-Curtis index 0.59 0.63 0.63 0.72 0.80 0.74 0.83 0.82 0.81 (0.24) (0.36) (0.29) (0.62) (0.73) (0.63) (0.74) (0.72) (0.72) Morisita’s index 0.86 0.86 0.89 0.88 0.81 0.88 0.96 0.97 0.97 (0.38) (0.43) (0.49) (0.76) (0.71) (0.77) (0.90) (0.92) (0.90) Jaccard’s index 0.54 0.53 0.52 0.51 0.51 0.38 0.47 0.54 0.41 (0.43) (0.42) (0.40) (0.44) (0.43) (0.31) (0.42) (0.49) (0.36) Notes: Numbers in parentheses are the critical values, where 95% of the generated communities had greater than or equal similarity to the observed community (i.e., an observed community with a lower value would be significantly changed). ‘‘PSI’’ is the percent similarity index. † Distribution is shown in panel A of Fig. 2. 1766 Ecological Applications Vol. 15, No. 5 JACOB SCHAEFER ET AL. the grand mean divided by the mean abundance during the given time interval. If the abundance for a time period was greater than the grand mean we used the reciprocal of the value. For example, if a species had a grand mean abundance of 50 individuals, and an abundance of 40 and 100 in the first two samples, the deviation of the first sample would be 1.25 (50/40), and 1.4 (70/50) for the first two samples combined. By definition, the deviation values asymptote at 1.0 when estimated means are equal to grand means. To quantify how sensitive our approach is to different levels of accuracy in forming reference samples, we ran simulations for each community using three different reference samples: (1) reference sample included only the abundances from the first sample, (2) reference sample included mean abundances from the first three samples, and (3) reference sample included mean abundance from the first six samples. These simulations were only conducted at the predicted level of 1.0 ⫻ CV. Programs for Monte Carlo simulations and associated calculations were written in the C programming language (see Supplement). Random walk simulations FIG. 3. Summary of statistically significant change (bars; left-hand vertical axes) and overall diversity (lines; right-hand vertical axes) for the nine communities examined. Open circles represent years in which no community change was detected; closed symbols represent years in which change was detected, with bars representing the number of metrics detecting change. Results shown are for expected level of variability (1.0 ⫻ CV) and one reference sample (first-year sample from each community). Annual, 4-year, and 20-year refer to experimental burn cycles in Konze prairie plots; LC, low change; MC, moderate change; HC, high change. To assess the sensitivity of our approach, 100 30-yr simulations for six stream fish communities were generated, each starting with 30 species (three species each with a mean abundance of 2, 4, 8, 16, 32, 64, and 100; two species each with a mean of 150, 200, and 250; one species each with a mean of 500, 750, and 1000). In the ‘‘no change’’ community, all species remained unchanged and abundance data at each time step were generated by a simple random walk. In the ‘‘two changed’’ community, the most abundant species declined over time from a mean of 1000 to a mean of two and the least abundant species increased from a mean of two to 1000 while the mean abundance of the other 28 species remain unchanged (random walk). The ‘‘six changed’’ and ‘‘12 changed’’ were similar except the three or six most abundant species decreased while the three or six least abundant increased. In the ‘‘six extinct’’ and ‘‘12 extinct’’ simulations the six and 12 least abundant species went extinct while the other 24 or 18 more abundant species remained unchanged. For each of the 600 simulations, the reference sample was the first generated (time 1) and the target sample the 30th (data in between were not used). Analyses were performed as described above using the CV–abundance relationship for stream fish. RESULTS samples across time, we evaluated the deviation of these estimates from the grand mean abundance across all sample years. For each time interval, we calculated the deviation from the grand mean for each species and plotted the mean deviation (⫾1 SE) for all species in that community. Deviation from the grand mean was defined as Community change Using the expected level of variation (1.0 ⫻ CV), the three LC communities showed significant differences between the reference samples and a target samples in 21.3% (49 out of 230) of the community ⫻ sample ⫻ metric combinations (Fig. 3). The MC and HC com- October 2005 ASSESSMENT OF COMMUNITY CHANGE 1767 deviation of estimated species abundance from the grand mean (mean for all stream fish species, Fig. 4a) and deviations for individual species ranged from 1.0to 77-fold differences from the grand mean. By averaging six samples to form our estimate the mean deviations for KP-LTER, NTL-LTER and stream fish were 2.9 (SE ⫽ 0.26), 2.1 (SE ⫽ 0.13), and 2.4 (SE ⫽ 0.23) times the grand mean, respectively, and the maximum deviation for a species was 18.3-fold (mean abundance over first 6 yr ⫽ 0.058, overall mean abundance ⫽ 1.063). For all three community types, the deviation of estimated mean abundances were highest with one to three years of data and dropped dramatically with four to six years of data. Increases in accuracy (decreased deviation) were not appreciable with more than six years of data (Fig. 4). Across all communities and metrics used, significant change was detected more often when reference samples were based on one years data (38.4% of target samples significantly changed) compared to three (33.2%) or six years data (25.9%, Fig. 5). Using quantitative indices, the number of communities detected as changed decreased as more samples were used to form the reference sample (one year, 50.9%; three years, 38.9%; and six years, 31.2%; Fig 5). The opposite trend was observed when using the qualitative Jaccard’s index as a metric (one year, 0.7%; three years, 16.0%; and six years, 10.0%; Fig 5), due to the accumulation of rare species over time as more samples were included in the reference sample. This effect was most obvious in the zooplankton data where a typical single year sample consisted of 36 species while the full data matrix for these communities averaged 70 species. FIG. 4. Accuracy of reference samples in estimating mean abundance for each of the three community types. Values on the x-axis represent the number of sample years used to calculate mean abundance. The y-axis represents the mean (⫾ SE ) deviation (estimated mean/actual mean) of the estimated mean from the grand mean for all species in each of the three community types. Sensitivity to level of CV Across all communities and metrics used, community change was detected in 78.4% of target samples using 0.5 ⫻ CV, 45.0% of target samples using 1.0 ⫻ munities showed significant differences in 52.6% and 60.4% of samples respectively (Fig. 3). Moreover, for the MC and HC communities, there was often agreement among metrics in detecting change, as compared to the LC communities. For example, community change was detected in 11 samples for the LC NTLLTER but only four times by more than two metrics. For the HC NTL-LTER data, community change was detected in 13 samples and always by four metrics. Accuracy of mean abundance in reference samples Estimates of mean species abundance over time asymptotically approached the grand mean as more samples were included in the reference sample. Using a single sample as an estimate of mean species abundance over time resulted in a maximum of a 6.8-fold FIG. 5. Sensitivity (total number of communities detected as significantly changed) of four similarity indices when species abundances in reference samples were estimated from one-, three-, and six-year means. Ecological Applications Vol. 15, No. 5 JACOB SCHAEFER ET AL. 1768 Random-walk simulations Of the quantitative metrics, Bray-Curtis and Morisita’s metrics were most sensitive to changes in abundance in the random-walk simulations (Table 3). All three of the quantitative metrics detected all simulated communities as changed when 12 of the 30 species either increased or decreased in abundance. Jaccard’s index was only effective at detecting extinctions, whereas the quantitative metrics did not consistently detect change even when the 12 least abundant species went extinct (Table 3). DISCUSSION FIG. 6. Sensitivity (percentage of communities detected as significantly changed) of four similarity indices and Euclidian distance in NMDS space when variance in species abundance from reference samples was based on 1.0, 0.5, and 2.0 times the coefficient of variation (CV). Species abundances from reference samples were based on the first-year sample. and 6.0% of target samples using 2.0 ⫻ CV (Fig. 6). In general, target samples found to be significantly changed at the 2.0 ⫻ CV level were drastically changed. Most (33 of 42) of the target samples detected as changed at the 2.0 ⫻ CV level occurred at one of the three HC sites. Conversely, communities where target samples were not detected as changed at the 0.5 ⫻ CV level would have been extremely stable. The Salt Fork of the Red River community was the most stable in this regard with only 33.7% (27 out of 80) of the target sample/index combinations indicating change at the 0.5 ⫻ CV level. CV , Sensitivity of individual metrics Combining all communities and CV levels, Euclidean distance in NMDS space, PSI and Bray-Curtis indices most often detected community change (61.2%, 51.7%, and 45.7%, respectively), followed by Morisita’s (40.9%) and Jaccard’s (16.29%; Fig. 6). PSI and BrayCurtis metrics were the most consistent in detecting change (agreeing in 85.4% of samples), whereas results from NMDS and Jaccard’s metrics differed the most often (28.0%, Table 2). In general, results among quantitative similarity indices were more similar than among NMDS and Jaccard’s index (Table 2). Based on our knowledge of the data sets used in our study, our approach provided an unbiased estimate of natural variability that was used to test for community change (significant difference between reference and target samples). In each of our community types, we detected change more often for HC communities than at MC or LC communities. Extensive data sets like those used in our analysis of LC, MC, and HC communities are helpful to demonstrate this technique; however such data are often unavailable for communities of interest. Our intentions were to develop a null model approach that can be used with a small number of reference and target samples. Because it is a randomization technique there is no loss of statistical power with these small sample sizes (Solow 1993, Sokal and Rolf 1994, Legendre and Legendre 1998). The fundamental assumption made with our approach is that species variability (CV) in abundance over time can be accurately predicted from abundance estimates from reference samples. Given the strength of the relationship between abundance and variation in abundance for the species in the three community types presented (stream fish, northern temperate lake zooplankton, and tallgrass prairie), this assumption appears to hold across a wide range of community types and is strong enough to generally assess change in the modeled communities. It is likely that similar relationships can be established with other community types, given the broad taxonomic range of organisms we examined in this study and relationships reported from other systems and taxonomic groups (Ford 1938, Grossman et al. 1990, Schoener and Spiller 1992, Winemiller 1996, Windberg 1997, Matthews 1998). It also is possible that the abundance CV in abundance relationships are TABLE 2. The percentage of times that pairs of similarity metrics both detected either change or no change in a sample (i.e., gave the same result) for all simulations with the first year as the reference and 1.0 ⫻ CV. Metric PSI Jaccard’s Bray-Curtis Morisita’s NMDS 84.5 28.9 74.9 64.8 (91.6) (53.0) (89.7) (78.2) PSI Jaccard’s Bray-Curtis 38.2 (59.4) 85.4 (91.6) 79.8 (85.3) 47.5 (58.8) 58.4 (60.0) 77.7 (84.6) Note: Numbers in parentheses indicate agreement at 0.5 ⫻ CV . Morisita’s ASSESSMENT OF COMMUNITY CHANGE October 2005 1769 TABLE 3. The percentage of simulated target samples (time step 30) from random-walk simulations that were detected as significantly changed from a reference sample (time step 1) for four metrics. Simulated community Metric PSI Jaccard’s Morisita’s Bray-Curtis No change Two changed Six changed 12 changed Six extinct 12 extinct 1 0 0 0 (0) (0) (0) (0) 2 0 6 15 (0) (0) (0) (0) 22 0 42 44 (0) (0) (0) (0) 100 0 100 100 Note: Model sensitivity was adjusted for 1.0 ⫻ (predicted parentheses) as described in Methods. transferable across systems. For example, the relationship between abundance and CV in abundance for Oklahoma stream fish may apply to other midwestern stream fish communities, thus, allowing a test for community change in any midwestern stream fish community with as little as one reference and one target sample. The abundance CV in abundance relationships for other community types could potentially be used in the same way, making this a useful technique for assessing change at sites that have not been intensely studied in the past. There are two important properties of this approach that should make it useful to ecologists: (1) it employs a Monte Carlo approach and (2) the model produces expected communities that can be analyzed with any index or metric of community similarity. We chose five commonly used indices and metrics to represent qualitative (Jaccard’s), quantitative (PSI, Bray-Curtis) and ordination (NMDS) approaches. It is clear that metric selection will affect the results of the analyses. For example, Jaccard’s index was much more sensitive to change for the NTL-LTER community because it had a large number of rare species that may not be detected in a given year. Conversely, PSI and Bray-Curtis will be much more sensitive to changes in the most abundant species in communities that are not as diverse and numerically dominated by a few species. These trends were also observed in analyses of the random-walk simulations (Table 3). We contend that our approach provides a framework with which an objective assessment of community change can be made and used with multiple metrics. Even IBI (Index of Biotic Integrity) scores can be generated for random communities to test the null hypothesis of no community change. Because of properties specific to each metric (Boyle et al. 1990), we recommend using our approach with a variety of qualitative and quantitative metrics and using the combined results to assess change in space or time. By using multiple metrics (e.g., qualitative vs. quantitative), one can assess what types of change the community has undergone (extinctions vs. significant change in abundances). At the same time, changing the expected level of variability (CV) in the model will yield information about the degree of change compared to background levels of change in training samples. (0) (0) (0) (14) CV ) 1 1 0 1 (0) (0) (0) (0) 1 100 1 1 and 2.0 ⫻ (predicted (0) (5) (0) (0) CV ) (in Reference sample accuracy and rare species The usefulness of this technique relies on the accuracy of the mean abundance estimates in the reference sample. In our analyses we found that averaging six samples through time provided accurate estimates of long-term mean abundance with diminishing returns when averaging more than six samples (Fig. 4). While it is tempting to average many samples into the reference sample, this will inherently bias the results by including more rare species than one would expect with one unit of sampling effort. The addition of more rare species will tend to bias the qualitative metrics more than quantitative or other metrics that can be adjusted to downweight rare species. This problem could be addressed by random selection of a portion of rare species, essentially standardizing the sampling effort represented in a reference sample. An alternate approach (one that is common used in community-level analyses; see Grossman 1985) would be to simply eliminate or downweight rare species. To examine what potential effect this would have on the model, we chose one community with a high proportion of rare species (annually burned KP-LTER community with 49 of 113 species not present in the reference sample) and performed simulations with rare species excluded. The results of the analysis were similar for the quantitative metrics (1993 sample not detected as changed by BrayCurtis, and 1995 sample not detected by as changed by PSI) and identical for the qualitative metrics (even though the mean Jaccard’s index in simulations increased more than any other, from 0.42 to 0.68). Overall, the presence or absence of rare species had little effect on metrics that are heavily influenced by abundance data and had limited effect on metrics that are influenced by presence/absence data. A second problem with including rare species (zero abundance in reference sample) is in how variability over time is calculated. With an abundance of zero in the reference sample and a predicted CV of 1.0 ⫻ CV, the SD is undefined. In our simulations, we calculated SD for these species as SD ⫽ CV ⫻ A where A was an arbitrarily chosen constant (1.0 for our communities). Thus, we included a low probability of occurrence and an abundance estimate for species that were not cap- 1770 JACOB SCHAEFER ET AL. tured in the reference sample, but were taken in one or more target samples. Changing this value to something other than 1.0 will have little effect at all on qualitative metrics but could drastically change results from quantitative metrics. We chose the arbitrary value of 1.0 because the abundance distributions generated with this value fit two of our three communities (KPLTER and NTL-LTER; data not shown). In these communities, raw abundance values were corrected for volume or area and mean abundance over time of 1.0 (as percent cover or number of organisms per 10 L) was reasonable for these rare species. This value may not have been optimal for the stream fishes since raw data were reported as number of individuals per collection. Thus, we assumed one individual of these rare species to be caught over 10 sampling periods. This arbitrary value is necessary if rare species are included in the analyses as only rare species need this constant to estimate SD in abundance. Another consideration is the length of the census period from which estimates of population variability are derived. For example, Pimm and Redfearn (1988) demonstrated that estimates of population variability increase with the length of the census period. It is therefore imperative that the temporal scale of the training samples match the temporal scale over which community change is assessed. For example, testing for community change over a three-year period with an abundance–CV curve based on 10 years of data might result in a test that is too conservative. In our data sets all abundances (or relative cover) were calculated as annual means and the CV–abundance curves generated from training samples covering the full length of each data set. One could therefore argue that our shorterterm comparisons (i.e., all but the last year at each site in Fig. 3) are overly conservative. This could explain why we observe more change in later years, but other studies of the same data sets (e.g., plant communities) using different techniques noted significant directional change (Collins 2000). In conclusion, a species mean abundance in reference samples can be used to predict how variable that species abundance should be over time. This relationship along with a reference sample is used to generate random communities one would expect in an undisturbed setting. We showed this null model can be used to test for temporal community change using a variety of metrics and common statistical techniques. We also demonstrated, through the use of simulated data sets, the sensitivity of different qualitative and quantitative indices of community change. The use of a variety of indices and parameterizing the model with different levels of species variability (i.e., 0.5, 1.0, and 2.0 ⫻ CV ) provides a context of community change to which target communities can be compared to determine the type and magnitude of change. Thus, with this flexible framework, ecologists can quantify and statistically test for community change. Such an assessment is not pos- Ecological Applications Vol. 15, No. 5 sible with current techniques. Although our data sets focused on temporal changes, this approach also can be used in a similar way to test for community change in space with a null model based on a relationship between abundance and variation in abundance in space. Identifying spatial and temporal changes in community structure is critical for assessing stability of communities subjected to anthropogenic and natural disturbances or other drivers of change. Moreover, information on background variability in community structure should prove useful to those making management decisions concerning disturbed systems that have not been extensively studied. ACKNOWLEDGMENTS We thank Tim Essington, William Matthews, Edie MarshMatthews, Kurt Shulz, Jim Rusak, Don Jackson, and two anonymous reviewers for assistance with this project at various stages. We thank SIUE Graduate School for funding. Konza Prairie Long-Term Ecological Research grant and the U.S. Geological Survey Gap Analysis Program provided support for K. B. Gido. The Knowledge Network for Biocomplexity project (DEB 99–80154) and National Center for Ecological Analysis and Synthesis, a Center funded by NSF (DEB-0072909), provided post-doctoral support for M. D. Smith. LITERATURE CITED Anscombe, F. J. 1948. The statistical analysis of insect counts based on the negative binomial distribution. Biometrics 5: 165–173. Bowman, M. F., K. M. Somers, and R. A. Reid. 2003. A simple method to evaluate whether a biological community has been influenced by anthropomorphic activity. Canadian Technical Report of Fisheries and Aquatic Sciences, Aquatic Toxicology Workshop. Minister of Public Works and Government Services, Ottawa, Ontario, Canada. Boyle, T. P., G. M. Smilie, J. C. Anderson, and D. R. Beeson. 1990. A sensitivity analysis of nine diversity and seven similarity indices. Research Journal Water Pollution Control Federation 62:749–762. Clarke, K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology 18:117–143. Clarke, K. R., and R. H. Green. 1988. Statistical design and analyses for a ‘‘biological effects’’ study. Marine Ecology Progressive Science 46:213–226. Clarke, K. R., and R. M. Warwick. 1994. Change in marine communities: an approach to statistical analysis and interpretation. Natural Environment Research Council, Plymouth, UK. Collins, S. L. 2000. Disturbance frequency and community stability in native tallgrass prairie. American Naturalist 155:311–325. Ebeling, A. W., S. J. Holbrook, and R. J. Schmitt. 1990. Temporally concordant structure of a fish assemblage: bound or determined? American Naturalist 135:63–73. Faith, D. P., P. R. Minchin, and L. Belbin. 1987. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69:57–68. Ford, J. 1938. Fluctuations in natural populations of Collembola and acarina. Part 2. Journal of Animal Ecology 7:350– 369. Gibson, D. J., and L. C. Hulbert. 1987. Effects of fire, topography and year-to-year climatic variation on species composition in tallgrass prairie. Vegetatio 72:175–185. Gotelli, N. J. 2001. Research frontiers in null model analysis. Global Ecology and Biogeography 10:337–343. October 2005 ASSESSMENT OF COMMUNITY CHANGE Gotelli, N. J., and G. L. Entsminger. 2003. Swap algorithms in null model analysis. Ecology 84:532–535. Grossman, G. D. 1982. Dynamics and organization of a rocky intertidal fish assemblage: the persistence and resilience of taxocene structure. American Naturalist 119:611–637. Grossman, G. D. 1985. Stochasticity and assemblage organization in an Indiana stream fish assemblage. American Naturalist 126:275–285. Grossman, G. D., J. F. Dowd, and M. Crawford. 1990. Assemblage stability in stream fishes: a review. Environmental Management 14:661–671. Jongman, R. H. G., C. J. F. Ter Braak, and O. F. R. Van Tongeren. 1995. Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, UK. Knapp, A. K., J. M. Briggs, D. C. Hartnett, and S. L. Collins. 1998. Grassland dynamics: long-term ecological research in tallgrass prairie. Oxford University Press, Oxford, UK. Legendre, P., and L. Legendre. 1998. Numerical ecology. Second edition. Elsevier, Amsterdam, The Netherlands. Magurran, A. E. 1988. Ecological diversity and measurement. Princeton University Press, Princeton, New Jersey, USA. Manley, B. F. J. 1991. Randomization and Monte Carlo methods in biology. Chapman and Hall, London, UK. Matthews, W. J. 1998. Patterns in freshwater fish ecology. Chapman and Hall, New York, New York, USA. McArdle, B. H., K. J. Gaston, and J. H. Lawton. 1990. Variation in the size of animal population: patterns, problems and artifacts. Journal of Animal Ecology 59:439–454. Micheli, F., K. L. Cottingham, J. Bascompte, O. N. Bjornstad, G. L. Eckert, J. M. Fischer, T. H. Keitt, B. E. Kendall, J. L. Klug, and J. A. Rusak. 1999. The dual nature of community variability. Oikos 85:161–169. Philippi, T. E., P. M. Dixon, and B. E. Taylor. 1998. Detecting trends in species composition. Ecological Applications 8: 300–308. Pigg, J., M. S. Coleman, J. Wright, R. Gibbs, K. Gido, and R. R. Lemmons. 1998. An ecological investigation of the ichthyofauna in the Deep Fork River, central Oklahoma: 1976 to 1996. Proceedings of the Oklahoma Academy of Sciences 78:67–110. Pimm, S. L., and A. Redfearn. 1988. The variability of population densities. Nature 334:613–614. 1771 Rice, J., and R. J. Belland. 1982. A simulation study of moss floras using Jaccard’s coefficient of similarity. Journal of Biogeography 9:411–419. Schoener, T. W., and D. A. Spiller. 1992. Is extinction rate related to temporal variability in population size? An empirical answer for orb spiders. American Naturalist 139: 1176–1207. Smith, E. P., K. W. Pontasch, and J. Cairns. 1990. Community similarity and the analysis of multispecies environmental data: a unified statistical approach. Water Research 24:507– 514. Smith, R. W. 2002. The use of random-model tolerance intervals in environmental monitoring and regulation. Journal of Agricultural Biological and Environmental Statistics 7: 74–94. Sokal, R. R., and F. J. Rolf. 1994. Biometry: the principles and practice of statistics in biological research. W. H. Freeman and Company, New York, New York, USA. Solow, A. R. 1993. A simple test for change in community structure. Journal of Animal Ecology 62:191–193. Spitzer, K., and J. Leps. 1988. Determinants of temporal variation in moth abundance. Oikos 53:31–36. Tilman, D. 1989. Ecological experimentation: strength and conceptual problems. Pages 136–157 in G. E. Likens, editor. Long-term studies in ecology: approaches and alternatives. Springer, New York, New York, USA. Warwick, R. M., and K. R. Clarke. 1991. A comparison of some methods for analyzing changes in benthic community structure. Journal of the Marine Benthological Association of the United Kingdom 71:225–244. Washington, H. G. 1984. Diversity, biotic and similarity indices: a review with special relevance to aquatic ecosystems. Water Research 18:653–694. Winemiller, K. O. 1996. Dynamic diversity in fish assemblages of tropical rivers. Pages 99–132 in M. L. Cody and J. A. Smallwood, editors. Long-term studies of vertebrate communities. Academic Press, San Diego, California, USA. Williamson, M. H. 1984. The measurement of population variability. Ecological Entomology 9:239–241. Windberg, L. A. 1997. Population trends and habitat association of rodents in Southern Texas. American Midland Naturalist 140:153–160. SUPPLEMENT The program Com Sim: a Monte Carlo community simulator designed to test for community change is available in ESA’s Electronic Data Archive: Ecological Archives A015-052-S1.