A genetic analysis of aminopeptidase and peroxidase isoenzymes in
advertisement

189
A genetic analysis of aminopeptidase and peroxidase isoenzymes in
Douglas-fir parent trees and seedling progeny
DONALD L. COPES
Forestry Sciences Laboratory, Pacific Northwest Forest and Range Experiment Station, United States Departmellf of Agriculture Forest Service, Corvallis, OR, U.S.A. 97331 Received June 23, 19781
Accepted December 4, 1978
CoPES, D. L. 1979. A genetic analysis of aminopeptidase and per'oxidase isoenzymes in Douglas­
fir parent trees and seedling progeny. Can. J. For. Res. 9: 189-192.
The genetic control of isoenzymes found in bark and vegetative buds was determined for one
peroxidase and one aminopeptidase locus. The segregation of alleles within full-sib progeny of
a 6 X 6 tree diallel fitted expected Mendelian ratios. Eight peroxidase isoenzymes existed,
but only those with relative mobilities of 0.44 and 0.49 showed allelism. Several of the other
six peroxidase bands gave a false impression of segregation.
CoPES, D. L. 1979. A genetic analysis of aminopeptidase and peroxidase isoenzymes in Douglas­
fir parent trees and seedling progeny. Can. J. For. Res. 9: 189-192.
Le contr6le genetique d'isoensymes trouvees dans l'ecorce et les bourgeons a ete determine
par un locus appartenant a Ia peroxydase et a une aminopeptidase. La segregation des alleles
pour une descendance biparentale d'un arbre 6 X 6 diallele confirma les rapports mendeliens.
De fait, huit isoensymes de Ia peroxydase etaient pn!sentes mais ce furent seulement celles
caracterisees par une mobilite relative de .0.44 et 0.49 qui manifesterent l'allelisme. De plus,
plusieurs des six autres bandes de peroxydase donnerent une mauvaise idee de Ia segregation.
[Traduit par le journal]
Electrophoretic techniques have been widely used
since 1968 to study variation in forest trees (see re­
view by Rudin 1976). Such techniques enabled forest
geneticists to study variation in what externally ap­
peared to be uniform populations. Knowledge has
been gained in the following areas: provenance and
geographic variation; lone, species, and hybrid
identification; climatic adaptation; developmental
changes; selection; disease resistance; differences be­
, tween dwarf and normal trees; mating system
analysis; and estimating levels of inbreeding.
In Douglas-fir (Pseudotsuga menziesii (Mirb.)
Franco) isoenzyme studies with starch or poly­
acrylamide gels have been reported for acid phospha­
tase (Baumeister 1975 ; Copes 1975), peroxidase (Juo
and Stotzky 1973; Mi.ihs 1974; Copes 1975), ribonu­
clease and phosphodiesterase (Merjnartowicz and
Bergmann 1977), aminopeptidase and esterase (Copes
1975; Juo and Stotzky 1973; Yang et a!. 1977),
glutamate dehydrogenase and catalase (Copes 1975),
and glutamate oxaloacetic transaminase (Yang eta!.
1977). In these studies isoenzymes from bark, needles,
bud meristems, embryos, or female gametophytes
were examined. Most enzymes in Douglas-fir have
multiple bands. Several enzyme systems have been
analyzed for genetic control (Miihs 1974), but no
studies have yet evaluated inheritance of isoenzymes
in progeny from controlled pollinations.
1Revised manuscript received November 24, 1978.
In this report, bark and vegetative bud tissues from
parent trees and their 2- and 3-year-old progeny were
studied. The inheritance of some of peroxidase and
aminopeptidase isoenzymes was determined.
Methods
In 1973, the six trees near Corvallis, Oregon, were control
pollinated in a 6 X 6 diallel design. Reciprocal crosses and
self-pollinations were omitted. Seeds were obtained from 14 of
the 15 families. In November 1975, about 30 seedlings of each
family were planted in the field near Monmouth, Oregon.
Tissues for electrophoresis were collected in December 1975,
November 1976, and December 1976. On each date, samples
were gathered from 400 progeny and 6 parent trees. A lateral
branch (10-12 em in length) was cut from the current year's
growth of each seedling and parent tree. Branches were stored
1 to 2 weeks in polyethylene bags at 0-2°C, Samples (50 to
75 mg) of the dome-shaped meristem of the terminal bud and
250-mg samples of living bark were used to study aminopepti­
dase and peroxidases, respectively. Bud and bark tissues were
macerated in one and three drops, respectively, of gel buffer
solution containing 59(; soluble polyvinylpyrrolidone (PVP-40,
molecular weight 40 000), absorbed on a 5 X 13 mm paper
wick (Whatman No. 1 chromatography paper), wrapped in
plastic, and stored at - 80°C until needed for electrophoresis.
Electrophoresis apparatus used was similar to that described
by Conkle (1972). Starch gel procedures used were identical to
those described by Copes and Beckwith (1977).
After electrophoresis, peroxidase was detected with o-dia­
nisidine (3,3'-dimethoxybenzidine) as substrate (Brewbaker
et a/. 1968) and aminopeptidase activity was detected with
L-leucyl-,8-naphthylamide as substrate (Scandalios 1969). !so­
enzymes from each gel were recorded in both diagrams and
photographs. Band positions were measured to the closest
CAN. J. FOR. RES. VOL. 9, 1979
190
1.00
-- -- -- -- -. - -Ano
��IJront-. -- -- ·
0.90
0.80
0.70
?;
::
:a
-o.12
· 0.60
h}
!
--a.49-o.44
0.40
:;�
.
'
:8
0.30
0.10
'J: 'J
Aminopeptidase
{
Peroxidase
.:... /1
·.·:.·'i\ t
-) ·
0.20
o.oo
{
....
...
......--o.ee
-o.ee,;r
0.60
0
E
iii
a:
-
J;
Origin elit--------- ------'"-""""'---'-"-"-'
2
1
3
4
6
6
Parent tree numbers
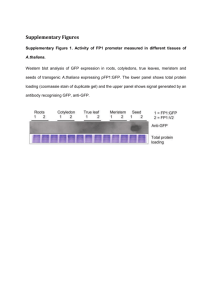
FIG. 1. Peroxidase and aminopeptidase zymograms of the six parent trees.
TABLE 1.
Family
(
X d')
1
2
3
4
4
X
X
X
X
X
Chi-squared goodness-of-fit test between observed and expected segregation ratios
among seedlings of full-sib families differing in peroxidase isoenzymes
Expected ratio
(Rro phenotypes)
0.44:0.44-0.49:0.49
4
4
4
5
6
0 :1:1
0:1:1
0 :1:1
0 :1 :I
0 :1:1
Totals
I
1
1
1
2
2
2
3
3
Cumulative
X 2
X 3
X5
X 6
X 3
X5
X 6
X5
X 6
Totals
Cumulative
2
x ,
2
x ,
4 df
I :2:1
1 :2:1
I :2:1
1:2:1
1 :2:1
I :2:1
1 :2:1
1:2:1
1:2:1
No. of observed seedlings/
No. of expected seedlings
(Rm phenotypes)
0 .44
0.44-o.49
0.49
(1)/0
0 /0
0 /0
0 /0
(1)/0
13/12.5
17/13.5
14/16.5
13/14
6/9
12/12.5
10 /13.5
19/16.5
15/14
12/9
0 /0
63/65.5
68/65,5
6/6.5
5/8.75
9/6.25
8/7
5/2.75
12/7 .25
11/8.25
10/7 .5
5/6.5
11/13
22/17.5
11/12.5
11/14
1/5.5
13/14.5
17 /16.5
15/15
13/13
9/6.5
8/8.75
5/6.25
9/7
5/2.75
4/7.25
5/8.25
5/7.5
8/6.5
71/60.75
16 df
114/121.5
Goodnessof-fit
test
pa
0 . 0 4•
1.82•
0.76 •
0 .14•
2.o o •
0.84
0 .18
0 . 38
0.71
0 .16
4.75
1.31d 2.83d 1.64d
1.36ll
7 . 36ll
4,72d
2.21d
1.67 d
0 . 69 d
0.31
0 .52
0.24
0.44
0 .51
O.G3
0.09
0.33
0.43
0 .7 1
23.7 9
0.0 9
58/60.75
ap, probability that x' will be exceeded, Null hypothesis should be rejected when value is less than 0.05,
Number In parentheses denotes aberrant zymograms that were deleted from x' analysis.
•Values were cnlculated using one degree of freedom. dValues were calculated using two degrees of freedom. millimetre and transformed into relative mobility (Rro) units
(the distance the isoenzyme travelled from the origin slit toward
the anode, divided by the distance the bromophenyl blue
marking dye moved from the origin slit toward the anode).
Segregation ratios observed in progeny were compared with
those expected from simple Mendelian inheritance. Tests of
homogeneity were made by the chi-square 'goodness-of-fit'
test.
Results
The gels stained for peroxidase revealed eight
isoenzymes but only the Rm 0.44 and 0.49 bands were
found suitable for genetic analyses. The Rm 0.44 and
0.49 bands were present in bark tissues from all three
collection times. They formed distinct bands which
COPES
TABLE 2.
Family
(9 X ci")
Chi-squared goodness-of-fit test between observed and expected segregation ratios
among seedlings of full-sib families differing in aminopeptidase isoenzymes
No. of observed seedlings/
No. of expected seedlings
(Rm phenotypes)
Expected ratio
(Rm phenotypes)
0.66:0.66-0.69:
0.66-0.72:0.69:
0.69-0.72:0.72
0.66
0:0:0:0:1:1
0:0:0:0:1:1
0:0:0:0:1:1
0:0:0:0:1:1
0:0:0:0:1:1
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
11/13.5
0/0
0/0
0/0
(1)/0
0/0
0/0
1X2
2X3
2X5
2X6
2X4
Totals
Cumulative
1 X4
3X4
4X5
4X6
Totals
Cumulative
1 X3
1 X5
1 X6
3X5
3X6
Totals
Cumulative
2
x ,
4 df
0:1:1:0:1:1
0:1:1:0:1:1
0:1:1:0:1:1
0:1:1:0:1:1
2
x ,
9 df
0:0:0:1:2:1
0:0:0:1:2:1
0:0:0:1:2:1
0:0:0:1:2:1
0:0:0:1:2:1
2
x ,
8 df
191
0.69-0.72
0.72
Goodnessof-fit
test
(4)/0b
0/0
(1)/0
0/0
0/0
7/13
6/5.5
16/14 . 5
13/16.5
0/0
9/13
5/5.5
13/14 .5
20/16.5
16/13.5
2.46°
0.09°
0.31°
1.49°
0.93°
0.12
0.76
0.58
0.22
0.34
11/13,5
0/0
42/49.5
63/53
6/6.5
7/8.25
7/7
5/4.5
8/6.5
6/8.25
7/7
5/4.5
(3)/0
(5)/0
(3)/0
(3)/0
7/6.5
12/8 .25
7/7
5/4.5
5/6.5
8/8.25
7/7
3/4.5
5 .28
0,77d
2 .52d
O.OOd
0.67d
0.26
0.86
0.47
1.00
0.88
0/0
25/26.25
26/26.25
0/0
31/26 .25
23/26 .25
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
0/0
(1)/0
0/0
0/0
0/0
7/8.5
6/6.25
7/6.75
8/7.5
9/6.5
18/17
10/12.5
12/13 .5
10/15
12/13
9/8.5
9/6.25
9/6.75
12/7.5
5/6.5
3.96
0.358
1.72•
1.93•
4 .408
1 .39•
0.91
0.84
0 .42
0.38
0.11
0 .50
0/0
0/0
0/0
37/35.75
62/71.5
44/35.75
9 .79
0.28
0.66-0.69
0.66-0.72
0.69
pa
ap, probability that x' will be exceeded. Null hypothesis should be rejected when value is less than 0.05,
bNu m bers In parentheses denote aberrant zymogra ms that were deleted fro m x' analysis.
<Values were calculated using one degree of freedom. dValues were calculated using three degrees of freedo m, •Values were calculated using two degrees of freedom .
stained faster and darker than other peroxidase
isoenzymes at Rm 0.27, 0.35, 0.66, 0.71, 0.77, and
0.80. These six bands were omitted from genetic
analysis because they were present in all trees (mono­
morphic) or were not repeatable.
Peroxidase zymograms of the six parent trees for
the Rm 0.44 and 0.49 bands are shown in Fig. I. Five
parents (I, 2, 3, 5, and 6) were heterozygous (Rm
0.44-0.49), but parent tree 4 was homozygous (Rm
0.49).
The progeny phenotypes revealed Rm 0.44 and 0.49
isoenzymes to be controlled by codominant alleles
located at one locus. Crosses between the five
heterozygous ( Rm 0.44-0.49) parents and the homo­
zygous (Rm 0.49) parent No. 4 yielded progeny in the
expected I: 1 Mendelian ratio (Table 1). No signifi­
cant x2 deviations were found. Crosses between the
four heterozygous (Rm 0.44-0.49) parents produced
progeny in the expected 1 :2: 1 Mendelian ratio
Table 1). Progeny from the 2 X 3 cross deviated
significantly at the 0.05 level, and it was a small
sample of only 11 seedlings.
Experimental error in the form of unexpected or
nonparental peroxidase phenotypes was detected in 2
of 373 trees. For example, progeny from a Rm
0.44-0.49 X Rm 0.49 cross should not have yielded
any trees with single-band Rm 0.44 phenotypes, yet 2
of the 133 seedlings from such crosses had aberrant
zymograms. The two trees were recorded in Table 1
but were not subjected to x2 analysis.
Four aminopeptidase isoenzymes were found in
extracts from buds at Rm 0.62, 0.66, 0.69, and 0.72.
The Rm 0.62 band was found in all six parent tr.r"'.'
and in all the progeny. This band appeared to b Jon '
trolled by a different locus from the other three bands.
Three bands (Rm 0.66, 0.69, and 0.72) were poly­
morphic when parents and progeny were considered.
The parental phenotypes are diagrammed in Fig. l.
Progeny evaluations of the Rm 0.66, 0.69, and 0.72
so enzymes indicate codo minant inheritance (Table
2). One or two of the bands, but not all three, were
present in each tree. Crosses of Rm 0.66-0.72 (parent
4) times Rm 0.69-0.72 (parents 1, 3, 5, 6) gave progeny
in the expected 1: 1: 1: 1 Mendelian ratio. Crosses of
Rm 0.66-0.72 (parent 4). or Rm 0.69-0.72 (parents 1,
3, 5, 6) times Rm 0.72-0.72 (parent 2) gave progeny in
the expected 1:1 ratio. Crosses of Rm 0.69-0.72
(parents
1, 3, 5, 6) times
Rm
0.69-0.72 did give
CAN. J. FOR. RES, VOL. 9, 1979
192
progeny in expected 1 :2:1 ratio. No chi-squares de­
viated significantly at the 0.05 leve1 from the expected
values.
Approximately 5% (21 of 395) of the progeny con­
tained some isoenzymes that could not be explained.
Aberrant isoenzymes were found in seedlings from
crosses of parent tree 4 (Rm 0.66-0.72) with Rm
0.69-0.72 trees (parents 1, 3, 5, 6). Fourteen progeny
from these crosses had a single Rm 0.69 band. Such
misclassified isoenzymes were considered experi­
mental errors and were noted in Table 2 but were not
subjected to chi-square analyses.
Discussion
· ·
Forest geneticists should proceed cautiously when
using isoenzyme data from diploid tissues to estimate
levels of heterozygosity in tree populations. Con­
sistent inheritance, persistent bands during develop­
ment, and freedom from environmental modification
are needed before heterozygosity can be accurately
estimated. The eight peroxidase isoenzymes detected
in this study illustrated this problem. Only the Rm
0.44 and 0.49 isoenzymes were suitable for genetic
analysis. The Rm 0.44 and 0.49 peroxidase isoenzymes
corresponded to the bands labeled 8 and 9 by Miihs
(1974). The Rm 0.77 and 0.80 bands gave a false
impression of segregation because of nongenetic
influences, because of developmental alteration of
gene expression, or because of variability in stain
visualization techniques. Classifying such isoenzymes
as segregating alleles would lead to erroneous
heterozygosity estimates.
The alleles of the peroxidase and the aminopep­
tidase loci were expressed codominantly. Both alleles
were equally dark stained in heterozygotes and all
bands were inherited as monomers. No 'hybrid'
bands appeared in the progeny. The recombination
.,f alleles in progeny corresponded to ratios expected
trom hypotheses of simple Mendelian inheritance.
Only 1 of the 14 families deviated significantly from
the expected ratio and that family consisted of a small
sample of only 11 seedlings. Inheritance of Rm 0.44
and 0.49 peroxidase isoenzymes in Douglas-fir closely
resembles the two alleles- one locus peroxidase
system reported in Chamaecyparis obtusa (Tajima
et a!. 1977).
The presence of nonparental isoenzymes in the
progeny may have been due to experimental error.
This inconsistency was small for peroxidase (0.5%)
but was much larger for aminopeptidase (5.0%).
Several factors may account for this difference. First,
aminopeptidase banC:s did not stain as darkly as
peroxidase bands. Very weak staining of one band in
a heterozygote could result from different substrate
specificities. Second, the aminopeptidase bands were
more difficult to accurately identify because of similar
Rm values. Other workers have noted similar experi­
mental error problems; i.e., Miiller (1976) found a
2% error in aminopeptidase data from a seed and
embryo study of Pinus virginiana, and Feret (1971)
reported a 5% experimental error in peroxidase
observations from Picea glauca needle tissues. Ade­
quate replication and uniform experimental pro­
cedure help minimize the nongenetic effects, but it is
important to realize they exist and are present in most
isoenzyme results. Further work on or use of poly­
phenol-inhibiting maceration solutions, such as those
reported by Kelley and Adams (1977) for juniper
leaves, might help reduce within-tree variation.
BAUMEISTER, VON G, 1975. Moglichkeiten der Friiherkennung
quantitativer Saatgutertragsleistungen bei Klonen von Pinus
silvestris L. in Samenplantagen. Silvae Genet. 24(5-6):
175---177.
BREWBAKER, J. L., M. D. UPADHYA, Y. MAKININ, and T.
MAcDoNALD. 1968. Isoenzyme polymorphism in flowering
plants. III. Gel electrophoretic methods and applications.
Physiol. Plant. 21: 930-940.
CONKLE, M. T. 1972. Analyzing genetic diversity in conifers ..
isozyme resolution by starch gel electrophoresis. U.S. Dep.
Agric. For. Serv. Res. Note PSW-264 .
CoPES, D. L. 1975. Isoenzyme study of dwarf and normal
Douglas-fir trees, Bot. Gaz. (Chicago), 136(4): 347-352.
COPES, D. L., and R. C. BECKWITH. 1977. Isoenzyme identifica­
tion of Picea glauca, P. sitc/rensis and P. lutzii populations.
Bot. Gaz. (Chicago), 138(4): 512-521.
FERET, P. P. 1971. Isoenzyme variation in Picea glauca
( Moench.) Voss seedlings. Silvae Genet. 20: 46-50.
Juo, P.-S, and G. STOTZKY. 1973. Electrophoretic analysis of
isozymes from seeds of Pinus, Abies, and Pseudotsuga. Can.
J, Bot. 51: 2201-2205.
KELLEY, W. A., and R. P. ADAMS. 1977. Preparation of ex­
tracts from juniper leaves for electrophoresis. Phyto
chemistry, 16: 513-516.
MEJNARTOWICZ, L., and F. BERGMANN. 1977. Variation and
genetics of ribonucleases and phosphodiesterases in conifer
seeds. Can. J. Bot. 55: 711-717.
MOHs, HAN s-J . 1974. Distinction of Dmtglas-fir provenances
using peroxidase-isoenzyme patterns of needles. Silvae
Genet. 2 3( 1 -3) : 71-76.
MOLLER, G. 1976. A simple method of estimating rates of self­
fertilization by analysing isozymes in tree seeds. Silvae
Genet. 25(1): 15-17.
RUDIN, D. 1976. Biochemical genetics and selection application
of isoenzymes in tree breeding. Int. Union For. Res. Organ.,
Proc. Jt. Meet. Adv. Generation Breed., Bordeaux, pp.
145-146.
SCANDALIOS, J. G. 1969, Genetic control of multiple molecular
forms of enzymes in plants: a review. Biochem. Genet. 3:
37-79,
TAJJMA, M., H. MIYAHIMA, and Y. MYAZAKI. 1977. Genetic
analysis for peroxidase isoenzymes in Cltamaecyparis
obwsa End!. (In Japanese with English summary.) J. Jpn.
For. Soc. 59: 173-177.
YANG, 1.-CH., T. M. CHING, and K. K. CHING, 1977. Isoen­
zyme variation of coastal Douglas-fir. I. A study of geo­
graphic variation in three enzyme systems. Silvae Genet.
26(1): 10-18.



![Anti-Thyroid Peroxidase antibody [1.B.10] ab31829 Product datasheet Overview Product name](http://s2.studylib.net/store/data/011964745_1-5f75fbef9c57765583cc9a2be1b15b4b-300x300.png)