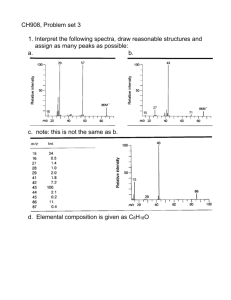
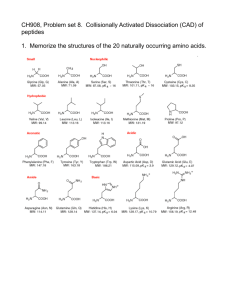
CH 908: Mass Spectrometry Lecture 8 Collisionally Activated Dissociation
advertisement
CH 908: Mass Spectrometry Lecture 8 Collisionally Activated Dissociation (of proteins and peptides) Prof. Peter B. O’Connor EI Mass Spectrum of an acetylated and reduced peptide Tandem Mass Spectrometry or MS/MS Isolation Fragmentation Isolation Fragmentation Benefits: 1.Extremely high MS/MS/MS, MS/MS or MS3 specificity 2.More structural information Limitations: 1.Isolation window 2.Fragmentation efficiency 3.Ion Losses Collisionally Activated Dissociation also called Collision Induced Dissociation (CID) N2 N2 + N2 N2 N2 N2 + N2 0 N2 • Ion’s smack into neutral gas • By far the most common molecules and break up MS/MS technique • Energy of the collision is controlled by changing the kinetic energy of the ion. • Fragments scatter radially • slow fragmentation method, deposits vibrational energy throughout the molecule prior to fragmentation. •SORI-CAD, ITMSn, Triple quad, TOF/TOF, etcetera Photo-Dissociation +* + + 0 hυ • Ion absorbs photon(s) and break •slow fragmentation method, deposits vibrational energy throughout the molecule prior to • Energy of the fragmentation fragmentation (depends on is controlled by changing the wavelength). photon’s wavelength. •IRMPD, UVPD, BIRD • No scattering, except for multiply charged ions Surface induced dissociation + 0 + • Ion smack into a surface, break, and rebound •slow fragmentation method, deposits vibrational energy throughout the molecule prior to • Energy of the fragmentation fragmentation. is controlled by changing the ion kinetic energy. •Ions are lost by neutralization at the surface (much better with • Fragments scatter radially perfluorinated surfaces) Outline: Collision models • Collision theory – – – – – – – Hard-sphere Soft-sphere Collision forces Langevin Cross-section Measuring cross-section Use of cross-sections – ion mobility Reactive collisions – ion molecule reactions – Internal energy deposition – Many, low energy collisions versus single high energy collisions • Peptide fragmentation nomenclature – Roepstorff – Biemann • • examples Preferential cleavage sites – Asp/glu – Pro •Structure of b,y ions •B2 ion •More examples •Breakdown diagrams •Proteins versus peptides •Oddball spectra – a/x ions in ubiquitin or CA Hard Sphere collision model 2 Area AB Valid for “high energy” collisions Langevin collision model 2 Area AB Ion Neutral z neutral 4 rIon Neutral 2 z=charge state r = ion-neutral distance α = polarizability Valid for “low energy” collisions Energy deposition During an ion-molecule collision, the fraction of kinetic energy that is lost by the ion is: 4mneutral mion 2 Ek E cos k 2 (mneutral mion ) Θ = scattering angle For the usual case of mion >> mneutral, and for the “worst case” scenario of a head-on collision (θ=0), this reduces to: 4mneutral Ek Ek mion Note: increasing the neutral’s mass, increases energy deposition This is the maximum amount of collision energy available, which will be distributed into translational, vibrational, and rotational modes. Internal Energy Conversion Typically, 20-50% of the ΔE is converted to internal vibrational energy. This ratio is a function of temperature, number of states, transition state energies of each reaction channel, etc. CAD/IRMPD/SID of Peptides and proteins ( ) i “standard” CAD spectrum of a peptide What’s in a sequence? Hemoglobin alpha chain: m/z 1529.7384 Roepstorff, P. and J. Fohlman (1984). "Proposal for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides." Biomed. Mass Spectrom. 11: 601. Amino Acid masses protonation sites Roepstorff, P. and J. Fohlman (1984). "Proposal for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides." Biomed. Mass Spectrom. 11: 601. Roepstorff, P. and J. Fohlman (1984). "Proposal for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides." Biomed. Mass Spectrom. 11: 601. Roepstorff nomenclature for peptide fragmentation. Roepstorff, P. and J. Fohlman (1984). "Proposal for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides." Biomed. Mass Spectrom. 11: 601. CAD, SID, IRMPD all produce b/y-type ions from peptides and proteins (primarily) Acylium structure n mass ai H 2O protons i 1 Protonated primary amine m mass ai H 2O protons in A Mobile Proton Theory of Peptide Fragmentation • The most stable protonated form may not be the fragmenting structure • Fragmentation (backbone) occurs due to the weakening of the amide bond, i.e. decrease of the bond order • Calculations showed that this will happen in the case of the protonation of the amide N • The more “mobile” (not localised) the proton, the more fragments in a MS/MS spectrum =>the more information from the spectrum Proton affinity Amino acid Proton affinity (kcal/mol) Proton affinity (eV’s) Lysine 110 4.8 Histidine 290 12.6 Arginine 315 13.7 Backbone amide ~40 1.7 Thus, the amino acids are protonated at Arginine first, then Histidine, then Lysine, then the backbone. Mobile proton model The Proline Effect in CAD/SID/IRMPD The Proline Effect in CAD/SID/IRMPD The Proline Effect in CAD/SID/IRMPD Selective Aspartic acid cleavage Tsaprailis, G., H. Nair, et al. (1999). "Influence of secondary structure on the fragmentation of protonated peptides." Journal of the American Chemical Society 121(22): 5142-5154. Tsaprailis, G., H. Nair, et al. (1999). "Influence of secondary structure on the fragmentation of protonated peptides." Journal of the American Chemical Society 121(22): 5142-5154. CAD/SID/IRMPD of Phosphopeptides Dehydroalanine Phosphoserine Serine CAD/SID/IRMPD of Phosphopeptides Phenylalanine Phosphotyrosine Tyrosine Comparison of CAD spectra on different instruments Tsaprailis, G., H. Nair, et al. (1999). "Influence of secondary structure on the fragmentation of protonated peptides." Journal of the American Chemical Society 121(22): 5142-5154. Relative frequency of Xxx-Zzz cleavage Xxx Zzz High energy CAD Immonium Ions: Glycans N-linked glycan O-linked glycan Self Assessment questions • What’s the main cleavage type for peptides/proteins under CAD/SID/IRMPD condition? Draw the structures of the fragments. • What additional fragment ions come from higher energy fragmentation? Draw the structures of the fragments. • Name two preferential cleavage points in peptide sequences. • What happens when a phosphoserine containing peptide undergoes CAD? • Memorize the structures of all 20 natural amino acids. (this is a very common viva question…) • Would a hard-sphere collision model or a langevin collision model yield a higher cross section for collision with Argon? CH908: Mass spectrometry Lecture 1 Fini… The Fragmenting Structure of a Protonated Peptide NH2 HN + NH H R1 O H N H2N O R2 N H OH O