Optimisation of preparative protein chromatography as part of whole bioprocess development
advertisement

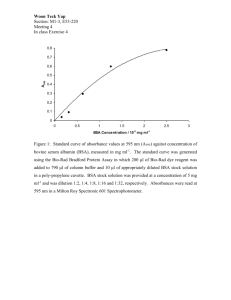
Optimisation of preparative protein chromatography as part of whole bioprocess development Kosma Jurlewicz Supervisor: Prof. Ajoy Velayudhan Group members: Dr. Spyridon Konstantinidis, Nicholas Field, Chris Wayne Background • Education: MSc, Eng. in Biotechnology (2005-2010, Technical University Wroclaw, Poland) – Specialisation: Biotechnological Processes • Work experience: 3 years experience in Industry (2010-2013, Polytherics [Abzena], London, UK) – Preparation, isolation and characterisation of new therapeutic protein candidates (e.g. PEGylation of proteins, conjugation of antibodies, ADC). • Interests: Hiking, climbing, travelling, art 2 Research aim • Develop a tool for rational optimisation of polishing step for challenging protein mixtures Mab processing from (Hanke & Ottens, 2014) based on (Shukla & Thömmes, 2010) Rationale: • Efficient removal of impurities in polishing step is crucial to achieve high purity of product and material throughput • There is a problem of HCP and product related impurities (glycosylation, deamination, phosphorylation and aggregation) • Lack of well characterised chromatographic profiles of complex protein systems under overloading conditions 3 Objectives • Find a model ternary protein system (Hemoglobin, Ovalbumin, BSA) • Generate preparative chromatograms • Optimise separation of model proteins (algorithms) • Gain more understanding in non-linear competitive chromatography • Apply this understanding to a real world mixtures from cell cultures 4 Mimicking product related impurities Protein profiles on Hb,Ova,BSA GE Capto Q column 1200 product Protein mix constrains: – Available (Sigma, etc.) – Cheap at g quantities (~ £ 20) – Representative size (> 50 kDa) – Binding to cation or anion exchanger – Binding with small amounts of salt present – Not fully separated / not fully coeluted on resin • • System: Hb, Ova, BSA Chromatography mode: anion exchange Hb Ova 1000 UV at 280 nm [mAU] • BSA weak 800 strong 600 400 200 0 4 6 8 10 Retention volume [CV] System: GE ÄKTA Run conditions: Column: HiScreen Capto Q 7.7 × 100 mm (4.7 mL) Binding: 50 mM Tris pH 9 + 50 mM NaCl Elution: 0 - 500 mM NaCl in 10 CV Protein load: 50 mg (10 mg protein/mL resin) 2 mL/min (260 cm/h) Hb loaded onto HiScreen Capto Q 5 Scheme of optimisation process 6 Preparative run and analysis Ternary mixture of Hb,Ova,BSA GE Capto Q column Recreated ternary mixture chromatogram GE Capto Q column 1200 12 product 800 strong weak Hb Ova 10 Quantification using analytical methods 600 HPLC / UV-VIS 400 200 Concentration [mg/mL] UV at 280 nm [mAU] 1000 2:6:2 BSA 8 6 4 2 0 0 4 6 8 10 12 Retention volume [CV] 14 4 6 8 10 12 Retention volume [CV] 14 As previously, apart from: Protein load: 120 mg (25 mg/mL resin) • Several conditions of model system tested, i.e ratio of weak/strong impurities to product • Fractionation and quantification of individual proteins allows to identify input parameters for optimisation • Mass balance was incorrect – suggesting inaccurate quantification 7 Challenges and relevance of the project Challenges • Accurate quantification of weak and strong impurities for the model system is challenging • Gaining high understanding of analytical problems and techniques before working with real bioprocess mixtures is needed Impact • Studying artificial mixtures, developing high throughput analysis methods and using fast optimisation algorithms will help improve relevant bioprocess separations • Rational optimisation of polishing step will reduce the cost of a bioprocess and ultimately benefit patients • Understanding of competition effects will contribute to the whole bioprocess development for even more challenging separations (mechanistic modelling) 8 Acknowledgments • Prof. Ajoy Velayudhan • Nicholas Field • Spiros Konstantinidis 9 Thank You for listening… …time for questions 10 Backup slide: 11 HPLC quantification method development HPLC analysis of Hb, Ova and BSA BIA CIMac™ QA column Hb Ova UV at 280 nm [mAU] 8 BSA 6 4 2 0 0 2 4 6 Retention time [min] 8 Conditions: 20 mM Tris pH 9 Elution: 0-350 mM NaCl, 5mM NaCl/ CV, 1 mL/min (280 cm/h) Protein load: 10 µg 10 • • • • Hb, Ova, BSA Loaded separately Range of concentrations Load plotted against AUC 12