'Translating massively parallel sequencing into clinical diagnostics.’ Children’s Hospital and
advertisement

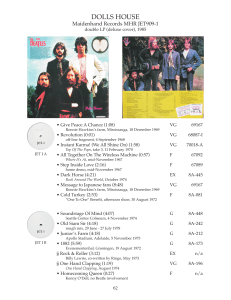
Women’s and Children’s Hospital ADELAIDE 'Translating massively parallel sequencing into clinical diagnostics.’ SA Pathology at the Women’s and Children’s Hospital and The University of Adelaide Adelaide, Australia Women’s and Children’s Hospital ADELAIDE MPS is revolutionising genetics & biology - >100 genes for various rare human disease discovered - de novo mutations (ID, ASD) identified, more to come? - understanding of the cancer, role of somatic mutation - diagnostic re-sequencing Women’s and Children’s Hospital ADELAIDE ESHG 2011 (Amsterdam, The Netherlands) May 28-31, 2011 H Brunner (Nijmegen, The Netherlands) Time to be in genetics! • • Capture only genes of interest Capture all and select only those of interest • Diagnostic problem = too many unclassified variants? • Why do diagnostic exome? • Cost effective (~ 2000 EURO/exome) • Clinical diagnosis not good enough as a guide for specific inquiry • Applicable to singletons • MPS is becoming a preference to clinical diagnostic testing • They aim for ~500 diagnostic exomes in 2011 Women’s and Children’s Hospital Child ADELAIDE 1/2011 10/2009 3/2010 3 /2010 AJHG A JH HG 10/2010 10/2010 0 4/2010 4 / 20 10 /2 4/2011 4 /2011 11/2010 1 1/2010 12/2010 12/2010 6/2011 Women’s and Children’s Hospital ADELAIDE Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 1/2011 753 pregnant women with high risk of trisomy 21 DNA molecules from 2.0–4.8mL of maternal plasm 6 bp ‘barcode’ indexing 86 had fetus with trisomy 21 (full karyotype based) Illumina GA II and IIx 753 pregnancies 79.1% 98.9% 91.9% 96.9% sensitivity 0.3 M reads /sample specificity positive predictive value negative predictive value 314 pregnancies 100% 97.9% 96.6% 100% sensitivity 2.3 M reads /sample specificity positive predictive value negative predictive value Women’s and Children’s Hospital ADELAIDE Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 1/2011 ‘Our data reveal that the main value of the maternal plasma DNA sequencing test is to rule out fetal trisomy 21.’ The false positive rates of the current screening programs = 5% Using MPS the trisomy 21 could be ruled out in 98% of those (5%) Leaving only 0.1% of women with referrals for amniocentesis or CVS Women’s and Children’s Hospital ADELAIDE X chromosome • 5% of the genome • 3% of the exome Women’s and Children’s Hospital ADELAIDE pter NLGN4 AP1S2 RSK2 ARX IL1RAPL1 TM4SF2 ZNF674 ZNF41 ZNF81 FTSJ1 PQBP1 SYP1 SHROOM4 JARID1C IQSEC2 HUWE1 FGD1 OPHN1 SLC16A2 DLG3 NLGN3 XNP/ATRX MAGT1 BRWD3 ZNF711 ACSL4 PAK3 AGTR2 CUL4B GRIA3 UPF3B ZDHHC9 SLC9A6 ARHGEF6 FMR2 (FRAXE) MECP2 RPL10 GDI1 SLC6A8 RAB39B 22.3 22.2 22.1 21.3 21.2 21.1 11.4 11.3 11.23 11.22 11.21 11.1 11 12 13 21.1 21.2 21.3 22.1 22.2 22.3 2 2 2 2 2 2 qter MID1, HCCS OFD1, FANCB CDKL5, NHS, SMS, PDHA1 GK DMD OTC BCOR, ATP6AP2 CASK, MAOA, NDP, PORCN, SYN1, PHF8 SMC1A, HADH2 ARHGEF 9 SLC16A2 MCT8, KIA2022, PGK1, ATP7A MED12 PCDH19 SRPX2 PLP TIMM8A NXF5 PRPS1 SIZN1A DCX LAMP2 UBE2A NDUFA1 OCRL FMR1 GPC3 IDS PHF6 ABCD1 HRPT L1CAM SOX3 AVPR2 FLNA IKBKG DKC1 Genetic heterogeneity of XLID • ~ 95 genes • ~ 14 new (unpublished) • >> families yet to be resolved Women’s and Children’s Hospital ADELAIDE X-exome re-sequencing EURO MRX/AU GOLD (V Kalscheuer and H Ropers) • Probands from 248 unsolved families • Agilet SureSelect; hybridisation based capture 47 657 baits; 7 591 exons • Sequenced using Illumine GA IIx • 96.8% regions covered by >3 reads • Average 4 novel missense variants/X chromosome • 99% sequence variants confirm by Sanger sequencing Women’s and Children’s Hospital ADELAIDE Alignment • eResearchSA supercomputer – “Corvus” (552 processors 4.46 trillion calculations per second) • Galaxy tool: http://galaxy.psu.edu/ • Variants reported at depth of 10 reads • Annotated using SeattleSeq – easy to use http://gvs.gs.washington.edu/SeattleSeqAnnotation/ Women’s and Children’s Hospital ADELAIDE Sequencing - Coverage • Illumina 65 bp single end reads • Two samples per lane ~8 x 106 reads per sample Women’s and Children’s Hospital ADELAIDE Chromosome-wide variation in coverage Women’s and Children’s Hospital ADELAIDE Chromosome-wide variation in coverage Women’s and Children’s Hospital ADELAIDE Example: Five X-chromosome ‘Exomes’ Mosaik Alignment Sample I II III IV V 57.9 82.8 60.9 42.9 91.1 Minimum 1 1 1 1 1 Maximum 2215 4434 2506 1830 3683 54 75 57 40 83 Mean Median # bases covered 2826330 2824649 2813718 2778055 2826976 # bases in bait region 3053381 3053381 3053381 3053381 3053381 % bases in bait covered 92.6% 92.5% 92.2% 91.0% 92.6% % bases in bait covered (10 fold) 86.2% 87.4% 85.2% 79.8% 86.9% S. Willis-Owen, unpublished. Women’s and Children’s Hospital ADELAIDE Highly Discrepant (HD) site Filtration Sample II III IV V 103,574 41,989 28,333 65,170 Within bait regions 92,190 36,893 25,130 59,366 Present in only 1 sample 54,599 9,677 5,819 24,315 210 153 133 149 No overlap with dbSNP130 63 17 26 23 Blat: best or only match to X 36 16 25 23 Missense 4 4 6 4 Predicted functional effect (any 1 of 4 tools) 2 1 2 3 All HD sites Proportion of reads matching ref allele <=15% S. Willis-Owen, unpublished. Women’s and Children’s Hospital ADELAIDE Zero coverage: Known ID Genes • 22/90 known (Gecz et al. TIGs 2009) XLID genes affected Gene ACSL4 AFF2 AP1S2 AR ARX BRWD3 DLG3 FGD1 G6PD HUWE1 IDS IQSEC2 OCRL PAK3 PDHA1 PGK1 SLC16A2 SLC6A8 SLC9A6 SOX3 SYN1 ZNF674 N bases 129 154 140 3 277 59 155 402 115 99 126 424 83 20 26 35 27 78 106 99 328 190 Women’s and Children’s Hospital ADELAIDE ARX gene Women’s and Children’s Hospital ADELAIDE X-exome re-sequencing - Results EURO MRX/AU GOLD (V Kalscheuer and H Ropers) 14 novel XLID genes identified 1 108 non-recurrent (novel) missense/STOP variants 366 non-recurrent (novel) silent variants ~ 90/248 families resolved – ~37% 61 families (25%) mutations in known XLID genes 29 families (12%) mutations in new XLID genes 38 families (15%) carry changes in candidate XLID genes 120 families (48%) unresolved Women’s and Children’s Hospital ADELAIDE X-exome re-sequencing - Results EURO MRX/AU GOLD (V Kalscheuer and H Ropers) 14 novel XLID genes identified 1 108 non-recurrent (novel) missense/STOP variants 366 non-recurrent (novel) silent variants ~ 90/248 families resolved – ~37% 61 families (25%) mutations in known XLID genes 29 families (12%) mutations in new XLID genes 38 families (15%) carry changes in candidate XLID genes 120 families (48%) unresolved Women’s and Children’s Hospital ADELAIDE Example 1: A small in/del detected by MPS (Family with X-linked Joubert syndrome with M Field and I Scheffer) I 1 1 II 1 * 2 2 2 3 4 3 5 6 7 8 9 III 1 2 * 3 4 5 6 7 * 8 IV 10 * Wt 1 * 2 3 * 4 5 6 7 2 ASD V 9 * 8 * 9 * Women’s and Children’s Hospital ADELAIDE pter qter Xp22.2 – Xp21.3 (hg18, chrX: 10,052,226- 27,452,338) Original New Mutation Codo A Codo A class n A n A • All HD sites 65,170 • 100% N Position % reads % reads Allele Chr read (bp) ref alt change s • Within bait regions 59,366 • 91.1% 27750326 chrX 48 2.10% 97.90% G/A 48006166 chrX 84 4.80% 95.20% T/G 48637316 chrX 22 15236403 9 chrX 18 4.50% 95.50% G/A TIMM17B Missense CGG R TGG W best X 5.60% 94.40% A/T TREX2 Missense CTG L CAG Q best X • Present in only 1 read 24,315 • 37.3% 149 23 23 4 Gene MAGEB1 0 Missense GTT V ATT SSX1 Blat I only X Missense TTC F TGC C best X • Proportion of reads matching ref allele <=15% • 0.23% • No overlap with dbSNP130 (1/2 bp features) • 0.035% • Best or only match to X (Blat) • 0.035% • Missense only • 0.006% p.230-235del IKMEAK of the OFD1 gene OFD1 NP_003602 200 NEYKREIEEQLRAEMCQKLKFFKDTEIAKIKMEAKKKYEKELTMFQNDFEKACQAKSEAL 260 OFD1 NP_003602 Del 200 NEYKREIEEQLRAEMCQKLKFFKDTEIAK------KKYEKELTMFQNDFEKACQAKSEAL 260 Women’s and Children’s Hospital ADELAIDE Example 2: mutation missed by large scale Sanger sequencing (with M Field and A Hackett, GOLD Newcastle) Women’s and Children’s Hospital ADELAIDE Sanger family #423 #423 ACSL4 Sanger sequencing Missed exon 13 (15) of ACSL4 and that is where the R654X mutation is! Women’s and Children’s Hospital ADELAIDE Example 3: Sanger and BAC aCGH negative (with M Field and GOLD NSW) 2 missense changes found of unknown significance A duplication involving DLG3 identified based on read depth Women’s and Children’s Hospital ADELAIDE Example 4: NO mutations found by Sanger or MPS (exomes) (with M Field and A Hackett, GOLD Newcastle) MRX3 studied since 1989 DXS304-Xq28; LODmax = 2.89 Women’s W omen’s s an and d Children’s Hospital C Ch Chi Chil d ’ss H dre dren ospi osp o os sp piita tal all a ADELAIDE Women’s and Children’s Hospital ADELAIDE Example 5: MRX12 22.3 22.2 22.1 21.3 21.2 21.1 OLD 11.4 11.3 11.23 11.22 11.21 11.1 (with M Field and A Hackett, GOLD Newcastle) LODmax = 3.01 11 12 13 21.1 21.2 21.3 22.1 22.2 22.3 23 24 NEW 25 26 27 28 Mutation in a novel XLID identified in the NEW linkage interval Women’s and Children’s Hospital ADELAIDE Example 6: IGOLD#586 family (with Julie McGaughran, Brisbane, QLD) Innocuous DNA variant found my MPS (1/4) 4 2 2 4 3 3 2 2 2 4 2 Expression profiling Pointed towards CCDC22 gene Women’s and Children’s Hospital ADELAIDE Summary • The MPS technology on its own will not address all your questions (find all variants) • Important to know what MPS can • do and what it can not do ALL or a PART (Whole Genome Sequencing or Targeted Enrichment and MPS)? • DNA variant annotation or functional effect interpretation • Coding variants vs non-coding variants • Missense variants vs silent variants • Unique variants vs recurrent variants • De novo variants vs inherited • SNVs vs small in/dels vs large CNVs Women’s and Children’s Hospital ADELAIDE Thoughts …. • MPS will not be the only technology in molecular diagnostic testing • MPS is likely going to be the first test (WG >> WE >> Targeted) • MPS (WGS) done only once & revised according to advancing knowledge (& annotation and tools for data mining) • Even if ~50% of MPS tests deliver the causal DNA variant $$ and effort well spent! Currently 1 exome ($1900) = Sanger sequencing of 1 gene (e.g. ARX). Women’s and Children’s Hospital ADELAIDE XLID NS-ARID >60 genes identified - 110 genes identified - - ~10-15% of ID - ~85-90% of ID ~ 500 genes causing ID Women’s and Children’s Hospital ADELAIDE How much rare variation? Women’s and Children’s Hospital ADELAIDE How much rare variation? Women’s and Children’s Hospital ADELAIDE How much rare variation? “The total somatic mutational load must be enormous. For example, the intestinal epithelium contains approximately 106 independent stem cells, each of which generates transient daughter cells every week or two. Thus, the intestinal epithelium of a 60-y-old is expected to harbor >109 independent mutations. This implies that, not far beyond the age of 60y, nearly every genomic site is likely to have acquired a mutation in at least one cell in this single organ.” Women’s and Children’s Hospital ADELAIDE M. Shaw, L. Hobson M. Corbett, L. Nguyen C. Shoubridge, A. Gardner L. Huang, S. Rujirabanjerd S. Willis-Owen, E. Douglas L. Jolly A. Hackett, M. Field, G. Turner I. Scheffer et al. H. Ropers, V. Kalscheuer Funding: Neurogenetics Research Program, Adelaide GOLD, Newcastle, AU Melbourne, AU Max Planck, Berlin NH&MRC Women’s and Children’s Hospital ADELAIDE Thank you for your attention.