Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011
advertisement

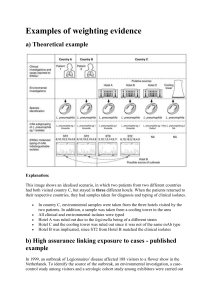
Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011 The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Grad, Y. H. et al. “Genomic Epidemiology of the Escherichia Coli O104:H4 Outbreaks in Europe, 2011.” Proceedings of the National Academy of Sciences 109.8 (2012): 3065–3070. Copyright ©2012 by the National Academy of Sciences As Published http://dx.doi.org/10.1073/pnas.1121491109 Publisher National Academy of Sciences Version Final published version Accessed Thu May 26 08:49:44 EDT 2016 Citable Link http://hdl.handle.net/1721.1/72563 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. Detailed Terms Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011 Yonatan H. Grada,b, Marc Lipsitchb,c, Michael Feldgardend, Harindra M. Arachchid, Gustavo C. Cerqueirad, Michael FitzGeraldd, Paul Godfreyd, Brian J. Haasd, Cheryl I. Murphyd, Carsten Russd, Sean Sykesd, Bruce J. Walkerd, Jennifer R. Wortmand, Sarah Youngd, Qiandong Zengd, Amr Abouelleild, James Bochicchiod, Sara Chauvind, Timothy DeSmetd, Sharvari Gujjad, Caryn McCowand, Anna Montmayeurd, Scott Steelmand, Jakob Frimodt-Møllere,f, Andreas M. Petersenf,g, Carsten Struvef, Karen A. Krogfeltf, Edouard Bingenh,i, François-Xavier Weillj, Eric S. Landerd,k,l,1, Chad Nusbaumd, Bruce W. Birrend, Deborah T. Hunga,d,m,n,2, and William P. Hanageb,1,2 a Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02115; bDepartment of Epidemiology, Center for Communicable Disease Dynamics, and cDepartment of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, MA 02115; dBroad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA 02142; eDepartment of Clinical Microbiology, Hillerød Sygehus, 3400 Hillerød, Denmark; fDepartment of Microbial Surveillance and Research, Statens Serum Institute, 2300 Copenhagen, Denmark; gDepartment of Gastroenterology, Hvidovre University Hospital, 2650 Hvidovre, Denmark; hLaboratoire Associé au Centre National de Référence des Escherichia coli et Shigella, Service de Microbiologie, Hôpital Robert Debré, Assistance Publique-Hôpitaux de Paris, 75019 Paris, France; iUniversité Paris-Diderot, Sorbonne Paris Cité, EA3105, 75505 Paris, France; jInstitut Pasteur, Unité des Bactéries Pathogènes Entériques, Centre National de Référence des Escherichia coli et Shigella, 75015 Paris, France; kDepartment of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139; lDepartment of Systems Biology, and mDepartment of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115; and nDepartment of Molecular Biology, Massachusetts General Hospital, Boston, MA 02115 The degree to which molecular epidemiology reveals information about the sources and transmission patterns of an outbreak depends on the resolution of the technology used and the samples studied. Isolates of Escherichia coli O104:H4 from the outbreak centered in Germany in May–July 2011, and the much smaller outbreak in southwest France in June 2011, were indistinguishable by standard tests. We report a molecular epidemiological analysis using multiplatform whole-genome sequencing and analysis of multiple isolates from the German and French outbreaks. Isolates from the German outbreak showed remarkably little diversity, with only two single nucleotide polymorphisms (SNPs) found in isolates from four individuals. Surprisingly, we found much greater diversity (19 SNPs) in isolates from seven individuals infected in the French outbreak. The German isolates form a clade within the more diverse French outbreak strains. Moreover, five isolates derived from a single infected individual from the French outbreak had extremely limited diversity. The striking difference in diversity between the German and French outbreak samples is consistent with several hypotheses, including a bottleneck that purged diversity in the German isolates, variation in mutation rates in the two E. coli outbreak populations, or uneven distribution of diversity in the seed populations that led to each outbreak. | food-borne outbreak Shiga toxin enterohemorrhagic E. coli | enteroaggregative E. coli | I n May–July 2011, two outbreaks of bloody diarrhea and hemolytic uremic syndrome (HUS) occurred in Europe: one centered in Germany (around 4,000 cases of bloody diarrhea, 850 cases of HUS and 50 deaths), and a much smaller outbreak in southwest France, near Bordeaux (15 cases of bloody diarrhea, 9 of which progressed to HUS) (1–4). Both outbreaks were caused by a strain of Shiga toxin-producing Escherichia coli of serotype O104:H4 (2, 5), which possesses a plasmid, pAA, characteristic of enteroaggregative E. coli, as well as a plasmid encoding an extended-spectrum β-lactamase (ESBL) (3). The proportion of patients infected with E. coli O104:H4 who develop complications, including HUS, is higher than seen in prior outbreaks (1, 6). The source of the outbreaks was epidemiologically linked to contaminated sprouts, and evidence indicates the outbreaks are connected to a 15,000-kg seed shipment from Egypt that arrived in Germany in December 2009. The majority of the seeds from the shipment (10,500 kg) was then sent to a German seed distributor, which supplied the implicated German sprout farm. Four hundred kilograms of the original seed shipment was sent to an English www.pnas.org/cgi/doi/10.1073/pnas.1121491109 seed distributor, which then repacked seeds into 50-g packets passed on to French garden stores (7). The seeds from a packet were then germinated into sprouts at a children’s community center, and the sprouts were served on June 8, 2011, leading to the French outbreak (2). Epidemiological investigations of outbreaks aim to combine various approaches to reconstruct in detail the chain of events that led to the outbreak. In principle, genetic information, such as the patterns of genetic diversity among isolates, can aid in tracking the origins and transmission of the pathogens. Genetic diversity can indicate how long the pathogenic lineage has been diversifying and shed light on when, where, and how this E. coli originated and entered the human food chain. In practice, such inferences require extensive and highly accurate genetic information. Even small error rates, which matter little for comparing an outbreak strain to historical isolates, could obscure genuine phylogenetic signal in comparing extremely closely related genomes from within an outbreak. Based on conventional molecular epidemiological characterization (including virulence gene content, serotyping, multilocus sequence typing, rep-PCR, pulsed-field gel electrophoresis, optical mapping, and antimicrobial susceptibility testing), the outbreak strains in Germany and France appear identical (2, 8) (see also Author contributions: Y.H.G., M.L., C.R., C.N., B.W.B., D.T.H., and W.P.H. designed research; Y.H.G., H.M.A., G.C.C., M. FitzGerald, P.G., B.J.H., C.R., S. Sykes, B.J.W., J.R.W., S.Y., Q.Z., A.A., J.B., S.C., T.D., S.G., C. McCowan, A.M., S. Steelman, E.B., F.-X.W., and C.N. performed research; Y.H.G., C.R., S. Sykes, B.J.W., J.R.W., S.Y., Q.Z., J.F.-M., A.M.P., C.S., K.A.K., E.B., and F.-X.W. contributed new reagents/analytic tools; Y.H.G., M.L., M. Feldgarden, M. FitzGerald, P.G., B.J.H., C.R., S. Sykes, B.J.W., J.R.W., S.Y., Q.Z., F.-X.W., E.S.L., C.N., B.W.B., D.T.H., and W.P.H. analyzed data; and Y.H.G., M.L., M. Feldgarden, C. Murphy, C.R., J.R.W., K.A.K., E.B., F.-X.W., E.S.L., C.N., B.W.B., D.T.H., and W.P.H. wrote the paper. The authors declare no conflict of interest. Freely available online through the PNAS open access option. Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. E. coli C227-11 AFRH01000000; E. coli C236-11 AFRI01000000; E. coli Ec04-8351 AFRL01000000; E. coli Ec11-3677 AFRM01000000; E. coli Ec09-7901 AFRK01000000; E. coli Ec11-4632-C1 AFVA01000000; E. coli Ec11-4632-C2 AFVB01000000; E. coli Ec11-4632-C3 AFVC01000000; E. coli Ec11-4632-C4 AFVD01000000; E. coli Ec11-4632-C5 AFVE01000000; E. coli Ec11-4404 AFUX01000000; E. coli Ec11-4522 AFUY01000000; E. coli Ec11-4623 AFUZ01000000; E. coli Ec11-5538 AFVF01000000; E. coli Ec11-5537 AFVG01000000; and E. coli Ec11-5536 AFVH01000000). 1 To whom correspondence may be addressed. E-mail: lander@broadinstitute.org or whanage@hsph.harvard.edu. 2 D.T.H. and W.P.H. contributed equally to this work. This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10. 1073/pnas.1121491109/-/DCSupplemental. PNAS | February 21, 2012 | vol. 109 | no. 8 | 3065–3070 MICROBIOLOGY Contributed by Eric S. Lander, December 29, 2011 (sent for review December 2, 2011) SI Materials and Methods). However, these approaches do not assess the full diversity among strains. A comprehensive strategy requires whole-genome sequencing with accurate resolution on the single nucleotide level and can be augmented by analysis of gene and plasmid content. Results We first performed whole-genome sequencing using the Illumina sequencing platform on four isolates from the outbreak centered in Germany (Table 1). Among these four isolates, we found only two SNPs relative to a published genome from the German outbreak, TY2482 (9): two of the isolates showed no differences relative to the reference, and two showed one SNP each (nucleotide positions 224851 and 1096014) (Table 2; see also SI Materials and Methods, Table S1, and Fig. S1). We independently confirmed the two SNPs by Sanger sequencing. As further validation of the sequence quality, we performed genome sequencing, assembly, and SNP calling of two of these isolates (C236-11 and C227-11), using an independent genome-sequencing technology (454 sequencing platform); this analysis found the same two SNPs and no additional ones (see SI Materials and Methods and Tables, S2, S3, and S4). Our observation of limited diversity in the German outbreak isolates is consistent with a recent report that found no SNPs in two independent isolates from the German outbreak (10). We then analyzed strains from the smaller French outbreak. We performed whole-genome sequencing on 11 isolates from seven patients, including five isolated simultaneously from a single patient (Table 1). Surprisingly, the diversity of the isolates from the French outbreak was considerably greater than that from the German outbreak (Table 2). We found 19 SNPs, all of which were validated by Sanger sequencing. The five isolates from the single host showed virtually no variation. Four isolates were identical, but the fifth lacked one SNP shared by the other four (Fig. 1A and Table 2). Technically, the low diversity within a single individual further confirms the sequencing quality. Scientifically, it suggests that infection may have involved a small inoculum [similar to the estimated low infectious dose of E. coli O157:H7 (11)], or that a small number of genotypes dominate within a host during an infection. A maximum-likelihood phylogeny of the outbreak isolates (Fig. 1A), rooted on historical E. coli O104:H4 isolates from 2004 and 2009 that we had also sequenced, showed that the limited diversity seen in the samples from the large German outbreak was nested within the greater diversity of French isolates. One SNP, at location 1568661, distinguishes the historical 2004 and 2009 isolates and all but two of the French isolates from the outbreak isolates from Germany. The most parsimonious explanation is that the isolates from the outbreak in Germany represent a subset of diversity seen in the French outbreak. We additionally placed the outbreak isolates into broader phylogenetic context using C227-11 as representative of the outbreak: historical E. coli O104: H4 isolates 55989 [isolated from an HIV-positive adult from the Central African Republic in the 1990s that, like the other isolates, is enteroaggregative, but, in contrast, is not Shiga toxin-producing (12)], 01–09591 [isolated from an individual in Germany in 2001 (13)], and the 2004 and 2009 isolates from individuals in France and a commensal E. coli genome E1167 (Fig. 1B). Although the historical E. coli O104:H4 isolates from 2001, 2004, and 2009 are related to this outbreak, they do not appear to be ancestral. To confirm that the diversity found in the French outbreak was absent in the German outbreak, we analyzed sequence data from eight additional German outbreak strains recently deposited in GenBank (GOS1, GOS2, H112180540, H112180541, H112180280, H112180282, H112180283, and LB226692). Although these genome sequences are not suitable for de novo SNP prediction using our approach (most lack quality scores), they can be evaluated for the presence of known SNPs. We found that none of these genomes contained any of the 19 SNPs seen in the French outbreak or the two identified in the German outbreak (see SI Materials and Methods for details), indicating that they share the same sequence as TY2482 at these sites. The identity of the SNPs suggests that they reflect recent diversification without evidence for either purifying or positive selection (14). Specifically, the SNPs are not biased toward protein-altering substitutions. Of the 21 SNPS, 3 (14.3%) SNPs are intergenic (in keeping with the range of 12.3–13.8% of the genome predicted to be intergenic) (Table S5). Of the 14 SNPs within coding regions, 4 (28.6%) are synonymous. We found that all German and French outbreak isolates contained the three plasmids, including pAA, the ESBL plasmid, and a much smaller third plasmid, all of which have been identified in other descriptions of the O104:H4 outbreak isolates (9, 10, 13, 15). Through synteny and ortholog analysis, we computationally predicted only one region of gene difference, a deletion in Ec115538, one of the French outbreak isolates (see SI Materials and Methods for details). We confirmed the absence of an 836-bp region in this genome by PCR analysis and note that it is adjacent to an insertion sequence. This deleted region includes three predicted genes and the 5′ end of a fourth predicted gene (SI Table 1. E. coli O104:H4 isolates sequenced and analyzed in this study Isolate name Date of symptoms Date of isolation Age Sex Clinical manifestations Outbreak Ec04-8351 Ec09-7901 Ec11-3677 Ec11-3798 C227-11 C236-11 Ec11-4404 Ec11-5536 Ec11-5537 Ec11-5538 Ec11-4632_C1 Ec11-4632_C2 Ec11-4632_C3 Ec11-4632_C4 Ec11-4632_C5 Ec11-4623 Ec11-4522 2004 2009 May 21, 2011 May 21, 2011 May 14, 2011 May 19, 2011 June 17, 2011 June 17, 2011 June 20, 2011 June 20, 2011 June 15, 2011 June 15, 2011 June 15, 2011 June 15, 2011 June 15, 2011 June 18, 2011 June 18, 2011 2004 2009 May 26, 2011 May 25, 2011 May 18, 2011 May 21, 2011 June 21, 2011 June 24, 2011 June 24, 2011 June 24, 2011 June 25, 2011 June 25, 2011 June 25, 2011 June 25, 2011 June 25, 2011 June 27, 2011 June 22, 2011 50 6 31 55 64 23 42 49 35 41 47 47 47 47 47 31 65 Male Male Female Male Female Male Male Female Male Female Female Female Female Female Female Female Female Unknown HUS Bloody diarrhea Bloody diarrhea Bloody diarrhea HUS HUS HUS HUS HUS HUS HUS HUS HUS HUS HUS HUS — — German German German German French French French French French French French French French French French 3066 | www.pnas.org/cgi/doi/10.1073/pnas.1121491109 Grad et al. B A 422387, 1546241 Ec11-5538 Ec11-4632.2 3621338 170476, 2029740 87 67 Ec11-4632.4 Ec11-4632.5 1256852, 2937308, 5226522 93 Ec11-4632.3 91 79 04-8351 09-7901 01-09591 Ec11-4632.1 95 Ec11-4623 C227-11 Isolates from single individual 55989 Ec11-5536 551216, 1262666, 2831655, 2932413 Ec11-4522 4114250, 4243327, 4660485, 4807228 2276417 1568661 Ec11-5537 Ec11-4404 1096014 65 224851 2252380 63 E1167 C236-11 C227-11 German outbreak isolates Ec11-3677 Ec11-3798 Ec09-7901 c EC04-8351 Historical isolates 0.05 0.002 Materials and Methods and Figs. S2–S4). We found no other evidence of gene gain or loss. Discussion In this study, we perform whole-genome sequencing of multiple isolates from the 2011 outbreaks of E. coli O104:H4 in France and Germany to identify differences among isolates that are indistinguishable by standard molecular epidemiological tools. We find that the isolates are all closely related, and that the German outbreak isolates have extremely limited diversity, whereas there is greater diversity among the isolates from the French outbreak. Several lines of evidence support our finding of extremely limited diversity among at least a majority of the German outbreak isolates. First, there is minimal diversity among the four independent isolates reported here (see Table 1 and Materials and Methods for description of the background of the isolates). Second, a previous analysis of two other isolates identified no SNPs between them (10). The chance of detecting a subpopulation that comprises 40% of the overall population using six randomly selected isolates is 95% [1 − (1 − 0.4)6 = 0.95]. Even in the absence of the two isolates from the independent analysis, the likelihood of detecting a subpopulation of 40% of the total population with four isolates is 87% [1 − (1 − 0.4)4 = 0.87]. Thus, our sample size is sufficient to detect, with high probability, variants present as a majority or large minority of all isolates. Third, eight isolates from the German outbreak with sequence in GenBank (GOS1, GOS2, H112180540, H112180541, H112180280, H112180282, H112180283, and LB226692) share identical sequence to TY2482 at the sites of each SNP position described in this study. Although it is impossible to exclude the possibility of unsampled diversity in the German outbreak, our findings argue that a majority of the population is extremely closely related. Using the framework of the trace-back epidemiology that links the two outbreaks to the 2009 shipment of fenugreek seeds, several hypotheses can explain the surprising findings that there is greater diversity of E. coli O104:H4 in the much smaller Grad et al. French outbreak than the German outbreak, and that the outbreak isolates from Germany appear to be nested within the diversity of the French outbreak (Fig. 2). One hypothesis is that the limited diversity reflects a stochastic bottleneck in at least the sampled part of the E. coli pathogen population in Germany compared with France. As we found no evidence for positive or purifying selection in the SNPs, the bottleneck we propose represents a random process that purged most of the diversity. The limited diversity observed within an individual suggests the hypothesis that the bottleneck in the German outbreak could represent contamination from a single infected human at the sprout farm in Germany. Consistent with this hypothesis, three employees were confirmed as early cases of E. coli O104:H4 infection, including two asymptomatic shedders, dating to around the time of the reported start of the outbreak in early May 2011 (16). In principle, the limited diversity in Germany could also result from partially successful measures to disinfect seeds or sprouts at the German sprout farm; however, it appears that no specific disinfection procedures were applied, apart from routine hygiene and cleaning of the sprout preparation area (16). Analysis of any isolates available from the earliest stages of the outbreak, including those from infected employees or sprouts, would allow for direct testing of these hypotheses. Broader sampling from the outbreak in Germany may help determine the extent to which the outbreak in Germany reflects contamination from a single individual, and whether there is evidence for subpopulations with additional diversity. A second hypothesis is that although substantial diversity was present in the original bacterial source population, it was unevenly distributed, with a more diverse population, perhaps reflecting heavier contamination, affecting seeds sent to France more than those sent to Germany. As a far greater amount of seeds (10,500 kg) went to the German distributor that supplied the establishment identified as the source of the German outbreak and only 400 kg went to the English distributor that supplied the 50-g seed packets believed to be the source of the PNAS | February 21, 2012 | vol. 109 | no. 8 | 3067 MICROBIOLOGY Fig. 1. (A) Bootstrap consensus maximum-likelihood phylogeny using the 21 SNPs, based on 500 bootstraps and rooted on the 2004 and 2009 isolates. No branch lengths are provided for the 2004 and 2009 isolates because this phylogeny is generated only from the 21 SNPs from the two outbreaks. The isolates associated with the German outbreak are C236-11, C227-11, Ec11-3677, and Ec11-3798. The 2004 and 2009 isolates are Ec04-8351 and Ec09-7901, respectively. The remaining isolates are associated with the French outbreak. Ec11-4632.1 to Ec11-4632.5 represent the five isolates from a single individual. The black numbers at nodes indicate bootstrap support. The maroon numbers along the branches indicate the locations, with respect to the TY2482 genome, of the SNPs that define each branch. (B) Bootstrap consensus maximum-likelihood phylogeny using SNPs derived from whole-genome alignment of assemblies of C227-11, 55989, 01–09591, Ec04-8351, Ec09-7901, and the commensal E. coli E1167, as described in Materials and Methods. The black numbers at nodes indicate bootstrap support. Table 2. SNPs identified within E. coli O104:H4 outbreak isolates SNP position 170476 224851 422387 551216 1096014 1256852 1262666 1546241 1568661 2029740 2252380 2276417 2831655 2932413 2937308 3621338 4114250 4243327 4660485 4807228 5226522 Gene/region Cyclic diguanylate phosphodiesterase domain-containing protein Calcium proton antiporter Primary amine oxidase HTH-type transcriptional regulator Aromatic amino acid transporter (tyrosine specific) Wzy NeuD family sugar O-acyltransferase Phosphatase yfbT dedA Intergenic between sulfite reductace hemoprotein β-component and phosphoadenosine phosphosulfate reductase L-asparaginase 2 Type 3 restriction enzyme/helicase OR PstII subunit sn-glycerol-3-phosphate Transport system permease ugpE Hypothetical protein di-haem Cytochrome c peroxidase family protein Intergenic: between soxR redox-sensitive transcriptional activator and yjcD putative permease 3-Isopropylmalate dehydratase large subunit Lysine decarboxylase 2 iniconductance mechanosensitive channel Intergenic: between hypothetical protein and citrate synthase Conserved hypothetical protein Isolates SNP* Substitution Ec11-4632 C1-C5 G→T Ser150Ile C227-11 Ec11-5538 Ec11-4522 C236-11 A→T C→T G→A C→T Glu366Val Ser703Leu Synonymous Synonymous Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 Ec11-4522 G→A A→T Arg361Gln Glu132Asp Ec11-5538 Ec04-8351; Ec09-7901; Ec11-4522; Ec11-4623; Ec11-4632 C1-C5; Ec11-5536; Ec11-5538 Ec11-4632 C1-C5 G→T G→T Ala48Ser Gly145Val C→A N/A Ec04-8351; Ec09-7901; Ec11-4404; Ec11-4522; Ec11-4623; Ec11-4632 C1-C5; Ec11-5536; Ec11-5537; Ec11-5538 Ec11-4404 T→C Leu271Pro A→C Asp393Ala Ec11-4522 C→A Cys99Stop Ec11-4522 Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 Ec11-4632 C2-C5 C→A A→C Arg73Ser Synonymous T→A N/A Ec11-5537 T→G Ile107Ser Ec11-5537 Ec11-5537 A→C C→A Lys367Gln Synonymous Ec11-5537 T→C N/A Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 A→T Asn2476Tyr SNP position is with reference to the TY2482 genome. N/A, not applicable, as SNP not in coding sequence. *SNP base differences are called with respect to the coding strand, rather than with respect to the Fasta sequence for TY2482. French outbreak (7), this hypothesis requires the low probability event that seeds with the higher diversity E. coli population happened to be in the smaller-sized shipments. Characterization of E. coli O104:H4 populations found on other seeds from this shipment may help to assess this hypothesis. To our knowledge, no such populations have yet been described. Finally, a third hypothesis is that the difference in diversity reflects unknown environmental or other constraints that influenced rates of accumulation of diversity once the bacteria arrived in each country. For example, it is possible that differences in sprouting conditions between the German sprout farm and the French community center could have led to differences in diversity. These differences in conditions include use of well-water at a temperature of 20 °C in the sprout farm in Germany (16), compared with tap water at ambient temperature (between 12 and 28 °C) in the French outbreak (2). Seeds in France were also germinated for about 1.5 d longer. Testing rates of accumulation of SNPs under various conditions may help to assess this possibility. Using next-generation sequencing methods, we have been able to reveal variation at a single nucleotide level within genome sequences from a point-source outbreak, all within a set of isolates 3068 | www.pnas.org/cgi/doi/10.1073/pnas.1121491109 that are identical by classic typing techniques. Highly accurate sequencing and SNP identification can overcome the noise from sequencing error and discern phylogenetic signal, which may, as in this case, depend on a small number of nucleotides. As demonstrated by the multiple independent sequencing efforts related to this E. coli O104:H4 outbreak (9, 10, 13, 15), and also epidemiological investigations of other infectious diseases (17–19), genomic epidemiology is likely to become the standard strategy in molecular epidemiology as the cost of sequencing continues to decline and technology becomes more widely accessible. The determination of genome sequence is already recognized as a vital part of investigating any new outbreak, to place the pathogen in context and gain insight into its origins and the basis of its pathogenicity. Together with other recent work (17–19), this study argues strongly for multiple genome sequences to understand patterns of transmission within an outbreak. Such analyses can already be conducted in a matter of days, and technological advances will only improve our ability to perform them in real time. The advantages of whole genome data include greater resolution than classic techniques for outbreak investigation, such as pulsed-field gel electrophoresis, and a body Grad et al. A B Data and inferences Physical separation hypothesis Seed population Seed population ? Possible unsampled diversity in German outbreak Diversity seen in the French outbreak C Diversity seen in the German outbreak D Variable mutation rate hypothesis Diversity seen in the German outbreak Bottleneck hypothesis Diversity in the seed population Seed population Mutation rate µ1 Diversity seen in the French outbreak Mutation rate µ2 Bottleneck Diversity seen in the French outbreak Diversity seen in the German outbreak Diversity seen in the French outbreak Fig. 2. Schematic of hypotheses to explain differences in E. coli SNP diversity seen in the French and German outbreaks. (A) At minimum, the contaminating population that gave rise to both the French and German outbreaks was polymorphic at location 1568661, and possibly other sites, indicating at least two types of genotypes in the original contaminating population (represented in green and blue). In the samples presented here, there is greater diversity in the French outbreak E. coli O104:H4 population than observed in the German outbreak population. Although the probability that our sample represents the majority of the German outbreak is high (see main text), unsampled diversity may exist in a minority of cases in the German outbreak. (B) By the physical separation hypothesis, there was uneven distribution of the diversity in the original contaminating E. coli O104:H4 population, with the 50-g seed packets that led to the French outbreak containing a greater degree of diversity than was present in the 75-kg of seed sent to the German sprout farm (7). (C) By the variable mutation rate hypothesis, the original seed population, comprising at least two genotypes, may have mutated more quickly along the route to the French outbreak because of environmental or other factors. (D) By the bottleneck hypothesis, the French outbreak diversity represents the original diversity present in the contaminated seeds. Either a subset or overlapping set of strains that led to the French outbreak were sent to the German sprout farm, followed by a bottleneck that restricted diversity in the German outbreak. A bottleneck could have taken place from the time of separation of the seeds from the original shipment to the German and English seed distributors through germination in the sprout facility. For discussion of factors favoring each hypothesis, see the main text. of data amenable to analysis with well-developed and understood phylogenetic methods. As this example demonstrates, the results of such analysis, combined with traditional epidemiology, can raise novel epidemiologic hypotheses and questions that are available only through sequencing of multiple isolates. Genome Sequencing. We used a multiplatform strategy, generating an average of 146-fold sequence coverage on the Illumina platform, supplemented with data from 454 and Pacific Bioscience platforms for specific analyses. For details of the sequencing methods and genome assembly, see SI Materials and Methods. Materials and Methods SNP Prediction and Validation. SNP calling was performed using our analysis pipeline [GATK v1.0.6011 (21)] based on alignments of paired-end read data (101 sequences from both ends of 180-bp insert fragments on the Illumina platform) to the TY2482 strain. Potential SNPs from the Illumina sequences were called by GATK Unified Genotyper (22), filtering the data according to the following parameters: >90% agreement among reads; at least five unambiguously mapped reads; no greater than 50% mapping ambiguity; insertions and deletions were ignored. Over 97% of the bases in the genome of each outbreak isolate fulfilled these criteria. Bases were identified that have the highest computational likelihood for calling a base as either agreement to the reference or a SNP. Only SNPs at locations where equally high-confidence calls could be made in all outbreak isolates were included in the analysis. At 54 sites, all outbreak and historical isolates showed the same sequence as each other but disagreed with the TY2482 reference genome; we did not identify these sites as SNPs and use them as discriminatory markers because they may represent errors in the reference sequence as opposed to true SNPs (Fig. S1 and Table S2). See SI Materials and Methods for details of 454-based genome sequencing and SNP validation and PCR-based validation. Strains Sequenced in This Study. Isolates include 4 linked to the outbreak centered in Germany, 11 from the outbreak in the Bordeaux area of France (of which 5 are from a single individual), and 2 2004 and 2009 Shiga toxin-producing O104:H4 isolates from France. The German outbreak isolates were linked by travel to Germany and timing of the cases. C227-11 derives from a 68-y-old woman originally from Hamburg, Germany, who was in Denmark when she fell ill; the isolate was obtained on May 18. Note that a genome sequence for this isolate was previously reported (15). To ensure consistency in our analyses, we independently sequenced this isolate and use the genome sequence we generated for the studies reported here. C236-11 was isolated from a 23-y-old man from Southern Denmark, which borders Germany, without confirmed travel to Germany; the isolate was obtained on May 21. Ec11-3677 derives from a 31-y-old German woman who had spent 2 wk in Northern Germany (May 5–21, 2011) and who was traveling in France at the time of illness on May 21. Ec113798 was isolated from a 55-y-old French man who traveled in Northern Germany between May 8 and 12, 2011, and had returned to France when he became ill on May 21. The French outbreak isolates (Ec11-4404, Ec11-4522, Ec11-4623, Ec11-4632_C1-C5, Ec11-5536, Ec11-5537, Ec11-5538) were collected from individuals in the same community near Bordeaux, all of whom were known to eat sprouts at a single event on June 8, 2011 (2). Ec04-8351 and Ec097901 were isolated from the stool of infected individuals in France in 2004 and 2009 and represent historical O104:H4 isolates (20) (Table 1). Grad et al. Phylogenetic Analysis. To study the phylogenetic relationship among the outbreak isolates, we created a single sequence for each isolate consisting of the genotype at the 21 SNP sites and used these data as input sequence to Mega (23). A maximum-likelihood tree was generated using the Kimura PNAS | February 21, 2012 | vol. 109 | no. 8 | 3069 MICROBIOLOGY Diversity seen in the German outbreak two-parameter model with 500 bootstraps and rooted on the branch leading to the 2004 and 2009 isolates. To study the relationship between the outbreak and historical isolates, we first aligned whole-genome assemblies of C227-11, 55989, 01–09591, 04–8351, 09–7901, and the commensal E. coli E1167 using progressiveMauve (24). We selected SNPs from this alignment that contain unambiguous bases for all isolates, are in regions that align, and have at least 90% agreement in a sliding 100-bp window around each SNP. These SNPs were used to generate a maximum-likelihood tree using the Kimura two-parameter model with 500 bootstraps and rooted on E1167. ACKNOWLEDGMENTS. We thank Flemming Scheutz from the World Health Organization Collaborating Centre for Reference and Research on Escherichia coli and Klebsiella, Department of Microbiological Surveillance and Research, Statens Serum Institut, Copenhagen, Denmark for the valuable contribution of strains and discussion; R. T. Chandler for his insights; all members of the Broad Biological Samples Platform and the Genome Sequencing Platform; Lynne Aftuck, Scott Anderson, Patrick Cahill, Marc Chevr- ette, Julio Diaz, Danielle Dionne, Sheli Dookran, Rachel Erlich, Stephanie Grandbois, Lisa Green, Andrew Hollinger, Laurie Holmes, Andrew Hoss, Edward Kelliher, Sharon Kim, Purnima Kompella, Tony LaCasse, Matthew Lee, Niall J. Lennon, Dana Robbins, Alyssa Rosenthal, Elizabeth Ryan, Brian Sogoloff, Todd Sparrow, Alvin Tam, Austin Tzou, Cole Walsh, and Emily Wheeler for sequencing and technical support; and Lucia Alvarado-Balderrama, Aaron Berlin, Gary Gearin, Sante Gnerre, Giles Hall, Alma Imavovic, David B. Jaffe, Annie Lui, Iain MacCallum, J. Pendexter Macdonald, Matthew Pearson, Margaret Priest, Dariusz Przbylski, Andrew Roberts, Filipe J. Ribeiro, Sakina Saif, Ted Sharpe, Terry Shea, Narmada Shenoy, and Shuangye Yin for computational and analysis support. This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN272200900018C (to B.W.B.), and the Infectious Disease Program of the Broad Institute; National Institute of General Medical Sciences Award U54GM088558 (to M.L. and W.P.H.); National Institutes of Allergy and Infectious Disease T32 Grant AI007061 (to Y.H.G.); Danish Council for Strategic Research Grant 09-063070 (to K.A.K.); and the Institut de Veille Sanitaire (F.-X.W. and E.B.). 1. Frank C, et al.; HUS Investigation Team (2011) Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. 2. Gault G, et al. (2011) Outbreak of haemolytic uraemic syndrome and bloody diarrhoea due to Escherichia coli O104:H4, south-west France, June 2011. Euro Surveill 16:pii: 19905. 3. Bielaszewska M, et al. (2011) Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect Dis 11:671–676. 4. Frank C, et al.; HUS investigation team (2011) Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Euro Surveill, 16: pii: 19878. 5. Scheutz F, et al. (2011) Characteristics of the enteroaggregative Shiga toxin/verotoxinproducing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill 16:pii: 19889. 6. Jansen A, Kielstein JT (2011) The new face of enterohaemorrhagic Escherichia coli infections. Euro Surveill 16:pii: 19898. 7. European Food Safety Authority (2011) Tracing seeds, in particular fenugreek (Trigonella foenum-graecum) seeds, in relation to the Shiga toxin-producing E. coli (STEC) O104:H4 2011 Outbreaks in Germany and France. Available at http://www.efsa. europa.eu/en/supporting/doc/176e.pdf. Accessed August 4, 2011. 8. Mariani-Kurkdjian P, Bingen E, Gault G, Jourdan-Da Silva N, Weill FX (2011) Escherichia coli O104:H4 south-west France, June 2011. Lancet Infect Dis 11:732–733. 9. Rohde H, et al.; E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium (2011) Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. 10. Brzuszkiewicz E, et al. (2011) Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 193: 883–891. 11. Armstrong GL, Hollingsworth J, Morris JG, Jr. (1996) Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev 18:29–51. 12. Touchon M, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. 13. Mellmann A, et al. (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6:e22751. 14. Rocha EP, et al. (2006) Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol 239:226–235. 15. Rasko DA, et al. (2011) Origins of the E. coli strain causing an outbreak of hemolyticuremic syndrome in Germany. N Engl J Med 365:709–717. 16. Bundesinstitut für Risikobewertung (2011) Relevance of sprouts and germ buds as well as seeds for sprout production in the current EHEC O104:H4 outbreak event in May and June 2011. Updated Opinion No. 23/2011 of BfR, 5 July 2011. Available at http://www.bfr.bund.de/cm/349/relevance_of_sprouts_and_germ_buds_as_well_as_ seeds_for_sprouts_production_in_the_current_ehec_o104_h4_outbreak_event_in_may_ and_june_2011.pdf. Accessed December 28, 2011. 17. Gardy JL, et al. (2011) Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. 18. Harris SR, et al. (2010) Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. 19. Rasko DA, et al. (2011) Bacillus anthracis comparative genome analysis in support of the Amerithrax investigation. Proc Natl Acad Sci USA 108:5027–5032. 20. Monecke S, et al. (2011) Presence of Enterohemorrhagic Escherichia coli ST678/O104: H4 in France prior to 2011. Appl Environ Microbiol 77:8784–8786. 21. McKenna A, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 22. DePristo MA, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. 23. Tamura K, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. 24. Darling AE, Mau B, Perna NT (2010) progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. 3070 | www.pnas.org/cgi/doi/10.1073/pnas.1121491109 Grad et al. Corrections and Retraction CORRECTIONS MICROBIOLOGY Correction for “Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011,” by Yonatan H. Grad, Marc Lipsitch, Michael Feldgarden, Harindra M. Arachchi, Gustavo C. Cerqueira, Michael FitzGerald, Paul Godfrey, Brian J. Haas, Cheryl I. Murphy, Carsten Russ, Sean Sykes, Bruce J. Walker, Jennifer R. Wortman, Sarah Young, Qiandong Zeng, Amr Abouelleil, James Bochicchio, Sara Chauvin, Timothy DeSmet, Sharvari Gujja, Caryn McCowan, Anna Montmayeur, Scott Steelman, Jakob Frimodt-Møller, Andreas M. Petersen, Carsten Struve, Karen A. Krogfelt, Edouard Bingen, François-Xavier Weill, Eric S. Lander, Chad Nusbaum, Bruce W. Birren, Deborah T. Hung, and William P. Hanage, which appeared in issue 8, February 21, 2012, of Proc Natl Acad Sci USA (109:3065–3070; first published February 6, 2012; 10.1073/pnas.1121491109). The authors note that on page 3066, right column, second full paragraph, lines 6–7, “Of the 14 SNPs within coding regions, 4 (28.6%) are synonymous” should instead appear as “Of the 14 SNPs within coding regions, 5 (36%) are synonymous.” Additionally, the authors note that Table 2 appeared incorrectly. The corrected table and its legend appear below. SNP position 170476 224851 422387 551216 1096014 1256852 1262666 1546241 1568661 2029740 2252380 2276417 2831655 2932413 2937308 3621338 4114250 4243327 4660485 4807228 5226522 Gene/region Cyclic diguanylate phosphodiesterase domain-containing protein Calcium proton antiporter Primary amine oxidase HTH-type transcriptional regulator Aromatic amino acid transporter (tyrosine specific) Wzy NeuD family sugar O-acyltransferase Phosphatase yfbT dedA Intergenic: between sulfite reductace hemoprotein β-component and phosphoadenosine phosphosulfate reductase L-asparaginase 2 Type 3 restriction enzyme/helicase OR PstII subunit sn-gluycerol-3-phosphate transport system permease ugpE Hypothetical protein di-haem cytochrome c peroxidase family protein Intergenic: between soxR redox-sensitive transcriptional activator and yjcD putative permease 3-isopropylmalate dehydratase large subunit Lysine decarboxylase 2 Miniconductance mechanosensitive channel Intergenic: between hypothetical protein and citrate synthase Conserved hypothetical protein Isolates SNP* Substitution Ec11-4632 C1-C5 G→T Ser150Ile C227-11 Ec11-5538 Ec11-4522 C236-11 A→T C→T G→A C→T Glu85Val Ser703Leu Synonymous Synonymous Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 Ec11-4522 Ec11-5538 Ec04-8351; Ec09-7901; Ec11-4522; Ec11-4623; Ec11-4632 C1-C5; Ec11-5536; Ec11-5538 Ec11-4632 C1-C5 G→A A→T G→T G→T Arg361Gln Glu132Asp Ala48Ser Gly145Val C→A N/A Ec04-8351; Ec09-7901; Ec11-4404; Ec11-4522; Ec11-4623; Ec11-4632 C1-C5; Ec11-5536; Ec11-5537; Ec11-5538 Ec11-4404 Ec11-4522 T→C Synonymous A→C C→A Asp393Ala Cys189Stop Ec11-4522 Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 C→A A→C Arg73Ser Synonymous Ec11-4632 C2-C5 T→A N/A Ec11-5537 Ec11-5537 Ec11-5537 Ec11-5537 T→G A→C C→A T→C Ile107Ser Lys367Gln Synonymous N/A Ec11-4623; Ec11-4632 C1-C5; Ec11-5536 A→T Asn2476Tyr SNPs identified within E. coli O104:H4 outbreak isolates. SNP position is with reference to the TY2482 genome. N/A, not applicable, as SNP not in coding sequence. *SNP base differences are called with respect to the coding strand, rather than with respect to the FASTA sequence for TY2482. www.pnas.org/cgi/doi/10.1073/pnas.1203955109 www.pnas.org PNAS | April 3, 2012 | vol. 109 | no. 14 | 5547–5548 CORRECTIONS AND RETRACTION Table 2. SNPs identified within E. coli O104:H4 outbreak isolates DEVELOPMENTAL BIOLOGY Correction for “Solute carrier family 3 member 2 (Slc3a2) controls yolk syncytial layer (YSL) formation by regulating microtubule networks in the zebrafish embryo,” by Aya Takesono, Julian Moger, Sumera Faroq, Emma Cartwright, Igor B. Dawid, Stephen W. Wilson, and Tetsuhiro Kudoh, which appeared in issue 9, February 28, 2012, of Proc Natl Acad Sci USA (109:3371–3376; first published February 13, 2012; 10.1073/ pnas.1200642109). The authors note that the author name Sumera Faroq should instead appear as Sumera Farooq. The corrected author line appears below. The online version has been corrected. Aya Takesono, Julian Moger, Sumera Farooq, Emma Cartwright, Igor B. Dawid, Stephen W. Wilson, and Tetsuhiro Kudoh www.pnas.org/cgi/doi/10.1073/pnas.1203335109 MEDICAL SCIENCES Correction for “Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes,” by Maria Kasper, Viljar Jaks, Alexandra Are, Åsa Bergström, Anja Schwäger, Nick Barker, and Rune Toftgård, which appeared in issue 10, March 8, 2011, of Proc Natl Acad Sci USA (108:4099–4104; first published February 14, 2011; 10.1073/pnas.1014489108). The authors note that Jessica Svärd and Stephan Teglund should be added to the author list between Anja Schwäger and Nick Barker. Jessica Svärd should be credited with contributing new reagents/analytical tools. Stephan Teglund should be credited with contributing new reagents/analytical tools. The corrected author and affiliation lines, and author contributions appear below. The online version has been corrected. Maria Kaspera,1, Viljar Jaksa,b,1, Alexandra Area, Åsa Bergströma, Anja Schwägera, Jessica Svärda, Stephan Teglunda, Nick Barkerc,2, and Rune Toftgårda,3 a Center for Biosciences and Department of Biosciences and Nutrition, Karolinska Institutet, Novum, 141 83 Huddinge, Sweden; bInstitute of Molecular and Cell Biology and Estonian Biocentre, University of Tartu, 51010 Tartu, Estonia; and cHubrecht Institute, Koninklijke Nederlandse Akademie van Wetenschappen and University Medical Center Utrecht, 3584CT Utrecht, The Netherlands Author contributions: M.K., V.J., and R.T. designed research; M.K., V.J., A.A., Å.B., and A.S. performed research; J.S., S.T., and N.B. contributed new reagents/analytic tools; M.K., V.J., A.A., Å.B., A.S., and R.T. analyzed data; and M.K., V.J., and R.T. wrote the paper. www.pnas.org/cgi/doi/10.1073/pnas.1203264109 RETRACTION IMMUNOLOGY Retraction for “The antibody zalutumumab inhibits epidermal growth factor receptor signaling by limiting intra- and intermolecular flexibility,” by Jeroen J. Lammerts van Bueren, Wim K. Bleeker, Annika Brännström, Anne von Euler, Magnus Jansson, Matthias Peipp, Tanja Schneider-Merck, Thomas Valerius, Jan G. J. van de Winkel, and Paul W. H. I. Parren, which appeared in issue 16, April 22, 2008, of Proc Natl Acad Sci USA (105:6109– 6114; first published April 21, 2008; 10.1073/pnas.0709477105). The undersigned authors wish to note the following: “In our study we employed Protein Tomography (PT), an electron tomography method commercialized by Sidec AB, for structural analysis of proteins. Following our publication, doubts were raised with respect to the validity of Sidec PT, and we therefore conducted an extensive validation study with Sidec AB to determine the fidelity of PT for structural analysis of therapeutic antibodies and antibody–antigen complexes in solution. In two independent double blind experiments, PT was found to be highly unreliable in distinguishing structural features of the molecules and complexes studied. First, approximately 90% of the identified protein density maps could not be interpreted due to complex morphology or low quality. Second, among the remaining objects, a high number of the protein images observed did not match with sample composition resulting in misinterpretation of sample identities. These disappointing results led us to reanalyze our previously acquired PT data in situ using a weighted backprojection reconstruction method with IMOD (1) rather than COMET (2), which also indicated our previous analysis to be unreliable. With the current PT methods, we were thus not able to validate tomograms neither obtained from vitrified proteins in solution nor aldehyde-fixed, stained cells. Therefore the undersigned authors no longer feel confident of the PT data presented in Figs. 3, 4, 5, and S4 of our study. The authors stand behind the supporting biochemical data provided in the manuscript and believe the proposed model to be plausible, the validity of which, however, should be addressed with other methods. We found it important to notify our colleagues of the specific technical flaws in our publication and apologize for any inconvenience caused. We hereby retract this manuscript.” Jeroen J. Lammerts van Bueren Wim K. Bleeker Annika Brännström Magnus Jansson Matthias Peipp Tanja Schneider-Merck Thomas Valerius Jan G. J. van de Winkel Paul W. H. I. Parren 1. Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of threedimensional image data using IMOD. J Struct Biol 116:71–76. 2. Skoglund U, Ofverstedt LG, Burnett RM, Bricogne G (1996) Maximum-entropy threedimensional reconstruction with deconvolution of the contrast transfer function: a test application with adenovirus. J Struct Biol 117:173–188. www.pnas.org/cgi/doi/10.1073/pnas.1203736109 5548 | www.pnas.org