MULTIVARIATE STATISTICAL PATTERN RECOGNITION OF CURIE-POINT PYROLYSIS-GAS CHROMATOGRAPHIC FINGERPRINTS
advertisement

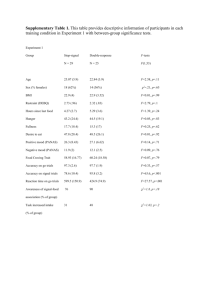
MULTIVARIATE STATISTICAL PATTERN RECOGNITION OF CURIE-POINT PYROLYSIS-GAS CHROMATOGRAPHIC FINGERPRINTS FROM RANGELAND SHRUBS D. N. Stevenson R. V. Valcarce G. G. Smith B.A. Haws E. D. McArthur B. L. Welch H. C. Stutz ABSTRACT The application of pattern recognition to the analysis of Py-GC data generally consists of two parts: unsupervised exploratory data analysis and supervised classification model development (Meglen 1988). Unsupervised exploratory data analysis detects outliers or abnormal measurements and provides information about the intrinsic data structure through classification. The goal of classification is to categorize a set of data as members of a class or classes without a prior or assumed knowledge of the data (Sharaf and others 1986; Wold and others 1984; Jerman-Blazic and others 1989). Unsupervised exploratory data analysis is an iterative routine that uses a variety of multivariate statistical methods such as cluster analysis, factor analysis, and principal component analysis, all of which are based on finding structural relationships or classifications among N-dimensional data (Meglen 1988; Tabachnick and Fidel11983). The three multivariate statistical programs used for exploratory data analysis in this study are: (a) LINK hierarchical cluster analysis (HCA); (b) MVSP principle component analysis (PCA); and (c) Fuzzy c-varieties pattern recognition (FCV). In general, these three techniques complement each other, and when used together provide a powerful tool for exploratory data analysis. Supervised classification model development is used to test the classification hypothesis determined in the exploratory data analysis phase by developing classification and prediction rules. These rules are used to predict class membership for new samples or to test the classification hypothesis by evaluating the performance of the rules on the data set (Sharaf and others 1986). Supervised classification model development relies heavily upon prior or assumed knowledge about class membership of the samples in the data set. Measurements or features of known samples are then used to construct a model that best represents the classification. Subsequent samples to be classified are compared with the classification model and assigned to an appropriate class (Knudson and others 1977; Meglen 1988). It is often desirable to reduce the number of features (pyrogram peaks) in the data set; this is accomplished The application of multivariate statistics to chemistry (chemometrics), using pattern recognition (PR) techniques, is shown to be a rapid and efficient method for the analysis of complex pyrolysis-gas chromatographic (Py-GC) data obtained from biomaterials. Results of two studies using various multivariate pattern recognition programs are presented. In one study, pyrograms obtained from accessions of big sagebrush (Artemisia tridentata) were correlated with differential palatability of the sagebrush to sheep. In the other study, Py-GC-PR was used to differentiate levels of ploidy in shadscale (Atriplex confertifolia). INTRODUCTION Pyrolysis-gas chromatographic (Py-GC) fingerprinting of complex biological materials has been shown to be a rapid and reliable chemotaxonomic technique (Soderstrom and Frisvad 1984; Torell and others 1989; Valcarce and Smith 1989a, 1989b). In pyrolysis, small amounts (usually micrograms) of directly sampled, underivitized material are fragmented by heating in the absence of oxygen. The resulting pyrolyzates are resolved by gas chromatography, producing a pyrogram. Pyrograms from biological samples, such as sagebrush and shadscale, are complex, and overall patterns of variation are not easily detected by visual examination. Multivariate pattern recognition techniques can be used to statistically evaluate and interpret·the data (Irwin 1982; Jurs 1986). Paper presented at the Symposium on Cheatgrass Invasion, Shrub DieOff, and Other Aspects of Shrub Biology and Management, Las Vegas, NV, April 5-7, 1989. D. N. Stevenson, R. V. Valcarce, and G. G. Smith are students and Professor Emeritus, Department of Chemistry and Biochemistry, Utah State University, Logan, UT 84322-0300; B. A. Haws is Professor Emeritus, Department of Biology, Utah State University, Logan, UT 84322-5305; E. D. McArthur and B. L. Welch are Project Leader and Research Plant Physiologist, Intermountain Research Station, Forest Service, U.S. Department of Agriculture, Shrub Sciences Laboratory, Provo, UT 84606; H. C. Stutz is Professor Emeritus, Department of Botany and Range Science, Brigham Young University, Provo, UT 84602. 325 This file was created by scanning the printed publication. Errors identified by the software have been corrected; however, some errors may remain. using an additional procedure known as feature selection, which ascertains the minimum number of variables (pyrogram peaks) necessary to correctly classify the training set samples (Duewer and Kowalski 1976; Sharaf and others 1986). In this study, feature selection was performed using the multivariate statistical program CART (Classification and Regression Trees). Applications of unsupervised exploratory data analysis and supervised classification and model development for the interpretation ofPy-GC data provide a powerful analytical tool for plant materials. The following two studies conducted on big sagebrush (Artemisia tridentata) and shadscale (Atriplex confertifolia) are presented to demonstrate the applicability of this technique for the analysis of genetically different but morphologically similar rangeland plants. In this study, pyrogram peaks that correlate with palatability of sagebrush to sheep were sought using supervised pattern recognition methods to classifY sagebrush pyrograms into clusters corresponding to three classes: low palatability (<25 percent), medium palatability (25 percent-75 percent), and high palatability (>75 percent). Shadscale Shadscale is also abundant in the Intermountain region of the western United States, from central Arizona and southwestern California to southern and eastern Montana. Although it is easily distinguished from all other species of saltbush (Atriplex sp.) populations are highly variable. Some variation may be attributed to environmental conditions, but the majority appears to be genetic, coming mostly from polyploidy and from introgression from other species (Stutz and Sanderson 1983). The chromosome numbers of shadscale plants are determined by cytological examination of meiotic cells in male flower buds. Collections for these studies can be made only during a few weeks each year when the plants are flowering. Being able to determine the ploidy level of shadscale at any time during the year would be useful. Since Py-GC-PR has been shown to be an effective method for discrimination of plant and insect materials (Soderstrom and Frisvad 1984; Torell and others 1989; Valcarce and Smith 1989a, 1989b), it was decided to attempt to characterize shadscale using these same methods. Preliminary Py-GC-PR studies were conducted using a limited data set of shadscale consisting of 16 plants representing eight locations and four chromosome races (table 2). Big Sagebrush Big sagebrush is among the most widespread shrub species as well as the most numerous single species in western North America (McArthur and others 1981; McArthur and Welch 1982). The big sagebrush complex is divided into three common subspecies: basin, Wyoming, and mountain big sagebrush (A t. ssp. tridentata, wyomingensis, and vaseyana) (McArthur and Plummer 1978; McArthur and Welch 1982), each consisting of various populations or accessions. Welch and others (1987) reported that domestic sheep showed differential preference for various accessions of big sagebrush (table 1). Differential palatability has applications in land rehabilitation, where less-preferred sagebrush could be used for revegetation in areas subject to overgrazing. Conversely, the establishment of preferred accessions of big sagebrush can provide winter forage, improving rangelands for domestic and wild animals (Behan and Welch 1986; Welch and others 1987). Table 1-Utilization of big sagebrush accessions by wintering sheep Table 2-Sample numbers, ploidy, identification numbers, and (Welch and others 19a7) Sample number 1-6 7-12 13-1a 19-24 25-30 31-36' 37-42 43-4a 49-54 55-60 Accession 1 Hobble Creek (v) Salina Canyon (v) Dove Creek (t) Petty Bish'ops Log (v) Clear Creek Canyon (v) Hobble Creek II (v) Clear Creek Canyon (t) Evanston (t) Milford (w) Evanston (w) location of shadscale samples used in this study Percent of current year's vegetative growth eaten ao.6 10.9 0.0 4a.3 21.6 ao.6 1.9 .5 a2.7 44.2 1 v =A. t. ssp. vaseyana, t =A. t. ssp. tridentata, w = A. t. ssp. wyomingensis. 326 Sample number Ploidy ID number 1-3 4-6 7-9 10-12 13-15 16-1a 19-21 22-24 26-27 2a-30 31-33 34-36 37-39 40-42 43-45 46-4a 2x 2x 2x 2x 2x 2x 4x 4x 4x 4x ax ax ax ax 10x 10x 79777 79777 a2246 a2246 a3244 a3244 a2272 a2272 a2261 a2261 a31aO a31aO a2239 a2239 a3133 a3133 Location Hardin, MT Hardin, MT Antelope Island Antelope Island Horse Canyon, UT Horse Canyon, UT Emery,UT Emery, UT Rock Springs, WY Rock Springs, WY Alkali Flats, OR Alkali Flats, OR Scipio, UT Scipio, UT Eskdale, UT Eskdale, UT EXPERIMENTAL represented by a pyrogram, can be considered as a N -dimensional data vector (Meglen 1988). Essentially, cluster analysis searches the distance matrix for the two data vectors with the smallest distance of separation. They are then treated as a single point, positioned at the center of gravity of the pair, and a new distance matrix is computed. This process continues with the number of groups reduced by one at each step, until all data vectors have been assigned to a single cluster (Dunn and Everitt 1982; Lavine 1988). A variety of methods to calculate the distance between a single point and a cluster, or between two clusters, are available (Romesburg 1984). Single linkage (SLINK), complete linkage (CLINK), and average linkage (UPGMA) between groups were the methods used in this study. Euclidean distances were used for generating the dissimilarity coefficient matrix. The results ofHCA are illustrated using a two-dimensional dendrogram that displays the multidimensional relationships among all samples (for example, fig. 1). Materials Big sagebrush samples, grown in uniform gardens established by the U.S. Department of Agriculture, Forest Service, Shrub Sciences Laboratory, Provo, UT, consisted of 10 accessions representing three different subspecies of big sagebrush (basin, mountain, and Wyoming big sagebrush) (Welch and others 1987). Shadscale samples taken from nursery-grown plants at Brigham Young University, Provo, UT, consisted ofleaves from 16 plants, representing eight different locations and four different chromosome races (2x, 4x, 8x, and lOx). Sample Preparation Each big sagebrush and shadscale sample was uniformly dried and ground to a fine powder. Fifteen milligrams of powder was suspended in 1.5 mL of spectralgrade methanol, and the resulting mixture was sonicated for 30 minutes. Portions (5-10 J..LL) of the sonicated mixtures were applied to 510 °C ferromagnetic pyrolysis wires and uniformly dried. MVSP Principal Component Analysis (PCA)PCA is a standard statistical technique used in numerical taxonomy (Dunn and Everitt 1982; Tabachnick and Fidell 1983). It reduces the dimensionality of multidimensional data but retains as much of the variation in the data as Pyrolysis-Gas Chromatography Analysis Each ferromagnetic sample wire was heated by induction, under helium, to 510 oc for 8 seconds using a F.O.M. XL Curie-point pyrolyzer. The resulting pyrolysis products were resolved on a 27-m (0.32-mm ID, 0.25-J..Lm film) Supelco SPB-5 fused-silica capillary column using a Hewlett-Packard 5880A gas chromatograph equipped with a flame ionization detector. Helium was used as the carrier gas, and the peak areas were determined with a Hewlett-Packard series 5880A level4 integrator. The pyrolyzer head was maintained at 85 °C, the gas chromatograph oven was heated from 50 °C to 200 oc at a rate of 5 °C/min, and the detector temperature was maintained at 200 °C. Dissimilarity Value 10 Evanston (t) 15 0.5'l'. Clear Creek Canyon (t) 1.9% L Dove Creek r (t) 0.0'1. L I l Data Processing Hobble Creek (vl 80.6'l'. The resulting pyrograms (retention time versus peak areas) were compiled into m X n data matrices, consisting of i = 1, 2, ... m samples andj = 1, 2, ... n features. Each data matrix was the starting point for further chemometric investigation by: LINK (HCA), MVSP (PCA), and FCVI?C-87 run on an IBM-AT-compatible equipped with a math coprocesser and an Orchid TurboPGA video card, and CART using a Digital Equipment Corporation VAX Model 8650. Data standardization was performed by normalizing each column of features in the data matrix to the sum of the values in the column. I Petty Bishop's Log (vl 48.3'1. I Clear Creek Canyon (v) 21.6'1. · '~ Salina Canyon (v) 10.9'1o L I Milford (w) 82.7'1. ---+---------....1 L Pattern Recognition Programs Figure 1-Dendrogram from hierarchical cluster analysis of sagebrush using single linkage (SLINK) between groups and Euclidean distance measure. LINK Hierarchical Cluster Analysis (HCA)-HCA uses techniques that search for unbiased natural groupings among samples inN-dimensional space. A sample, 327 20 25 possible. This enables direct examination of relative positions of the data points (pyrograms) in the highdimensional space. This is accomplished by transforming the original variables (pyrogram peaks) into a set of new uncorrelated variables known as principal components (PC's). The resulting PC's are linear combinations of the original variables and are arranged in order of decreasing variance, relative to the variation originally present in the data (Tabachnick and Fidel11983). If the axes of the first two or three principal components account for most of the variation, plots can be generated (PC1 vs. PC2 or PC3) to represent the relative positions of the data points in the high-dimensional space. among the three palatability classes in table 3. A variety of other methods of supervised classification are available, including discriminant analysis or soft independent modeling of class analogy (SIMCA). In this study, supervised classification was performed by normalizing the three palatability classes separately. Figure 2 shows the threedimensional "false color" plot of the data structure determined using this supervised procedure. An additional weighting feature relating the palatability of the sagebrush to sheep was added to the data set. This resulted in a "forced clustering" where the palatability features were unnormalized (values from 0-100), and the pyrogram peak areas were normalized (global average value = 1.00), producing the three classes of sagebrush samples: low palatability (30 samples), medium palatability (12 samples), and high palatability (18 samples). FCV allows the samples to have a shared class membership. This was particularly Fuzzy c-Varieties Pattern Recognition (FCV)FCV pattern recognition consists of two parts: multi class principal component modeling (MPCM) and false-color data imaging. The objective of the MPCM algorithm is to obtain disjoint principal component models of the classes within the data (Gunderson 1984; Jacobsen and Gunderson 1987). Multiclass principal component modeling is an unsupervised agglomerative method that determines the membership of a sample class within a preselected number of classes using a variance-based optimization routine (Vogt and others'1989). In the MPCM algorithm, each sample data vector plays a weighted role in defining each class represented by the data. The result is a membership matrix, containing membership values for each sample data vector in each class. The output of the MPCM algorithms, a set of principal components, one per class, and a membership coefficient matrix (membership values), can be used for cluster analysis, classification of new samples, and feature selection. False-color data imaging (Gunderson and others 1988), a plotting subroutine, makes it possible to evaluate model validity using three-dimensional "false color" images and is useful for evaluating the results of the algorithm. Table 3-Ciassification of big sagebrush accessions according to palatability Palatability level Accession Subspecies Percent used High Hobble Creek Milford A t. ssp. vaseyana A t. ssp. wyomingensis 80.6 82.7 Medium Petty Bishop's Log Evanston A t. ssp. vaseyana A t. ssp. wyomingensis 48.3 44.2 Low Evanston Clear Creek Canyon Dove Creek Clear Creek Canyon Salina Canyon A A A A A Classification and Regression Trees (CART)CART classifies samples according to tree-structured rules (Breiman and Friedman 1984). The classification is performed according to a probability model. On a test set, all features are used to construct a large tree, which is then pruned to the minimal tree necessary to perform the classification. A cross-validation procedure can be used to determine the significance of the features or variables. The success or significance of the classification is expressed as a misclassification rate. CART can be used to develop a test set and to analyze unknowns. t. ssp. t. ssp. t. ssp. t. ssp. t. ssp. tridentata tridentata tridentata vaseyana vaseyana 0.5 1.9 .0 21.6 10.9 y Moderate Palatability &~~~-~ RESULTS z Big Sagebrush Using the single-linkage (SLINK) clustering method, HCA was applied to the big sagebrush data set where the clustering was found to follow a hierarchical pattern (fig. 1). Figure 1 shows that classes representing different sagebrush accessions were detected, although no relationship with palatability could be established. However, using the supervised approach of the FCVMPCM algorithm, a good discrimination was obtained Figure 2-Three-dimensional false color data image plot of the three sagebrush palatability FCV classes determined using supervised classification. 328 X useful in the construction of the palatability classes because it allowed samples with 20 percent palatability to sheep to be partly in the "low" class and partly in the "medium" class. Of particular interest are the pyrogram peaks that show a stepwise decrease or increase relative to low-, medium-, and high-palatability classes(* in fig. 3). The FCV center values (average value of each class center in 37 dimensional spaces) are graphed in figure 3. A slight increase with increasing palatability is observed for features 2 and 27, and a more pronounced increase for features 20, 22, and 35. A stepwise decrease is observed for features 7, 15, and 31. Figure 3 also reveals that several other peaks have much higher discriminating power than the peaks mentioned, such as, 4, 9, 13, 14, 17, 19, 26, and 36 for the low class, and 12, 18, 23, 24, 25, and 32 for the medium class. Since the different classes contain different accessions of sagebrush, these peaks may represent chemical compounds useful in numerical taxonomy, but not necessarily correlated with palatability. CART is designed for classification (supervised data analysis); therefore, an exploratory data analysis cannot be performed. CART was applied to the data set in which the palatability value was added as a class variable (low = 1, medium= 2, high= 3), and the chemical data were used to classify the samples into the three classes. The resulting tree (fig. 4) consists of three nodes where samples are split into two groups according to the value of a certain feature. At node 1 of the tree, the samples were classified according to feature 20 S 0.905. Thirty-six samples were found with feature 20 S 0.905, as shown in figure 4. These samples consist of both high-palatability class samples and medium-palatability class samples. Node 2 in the classification tree was used to classify the 36 samples according to feature 32 S 4.51. At this node, 30 samples were found with feature 32 S 4.51, and were assigned to the high-palatability class; six samples were found with feature 32 ~ 4.51 and were assigned to the mediumpalatability class. The 24 samples at node 1 with feature 20 ~ 0.905 consisted ofboth medium- and low-palatability classes. Node 3 was used to split the 24 samples into medium- or low-palatability classes according to feature 25 ~ 4.57. In addition to the construction of the classification tree, CART also evaluates the importance of the different features. Some features compete with those used in the tree. A ranking according to importance may, therefore, give high priority to features not used in the classification step. Table 4 shows the relative importance of the different features determined by CART. Feature 20 was highest; all other features that show a stepwise increase with increasing palatability have an importance greater than 54 percent. Figure 5 shows the 20 pyrogram peaks best used for distinguishing palatability. 5 • 4 - 3 - Low Palatability 0 Med Palatability IJ High Palatability * 2 * * 1 • > * * * : * : : : 0 ~ . ~ l . .I : . J ~ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 Pyrogram Peak Figure 3-Center values for the three sagebrush palatability FCV classes determined using supervised classification. The .. above the bars indicates the pyrogram peaks with a stepwise increase or decrease relative to the low-, medium-, and high-palatability classes. 329 Table 4-Relative importance of features according to CART Feature number Relative Importance Feature 20 ~ 0.905 v 3( rf \s\4 r-~~--. ~~----~ Medium Palatability Low Palatability High Palatability 20 24 25 18 3 23 22 27 2 1 6 29 35 Feature 25 ~ 4.57 '6 Relative importance Percent 36/ '24 Feature 32 ~ 4.51 Feature number Percent 100 75 75 75 74 71 68 65 63 60 59 55 54 48 48 48 47 47 47 47 42 41 39 28 26 <25 12 32 21 4 13 19 26 28 5 34 10 31 Remaining ...... ·.- Figure 4-Ciassification according to CART. At each of the nodes, one feature is used to split the data set into two subsets. Final classification of sagebrush samples according to palatability is: high, 30 samples; medium, 12 samples; and low, 18 samples. 6 II Low Palatability ~ Med Palatability 5 - IIJ High Palatability 4 . 4 - 3 - 14 3 15 v v v v 6 v 17 2 2 19 18 1 7 1 . 10 9 11 5 v v v v v v 0 1 2 3 1/ 4 6 v v v v v v v._ 12 13 v 20 16 v v v - v ,.... v v v , - • 18 19 20 ~-I v v , v ~/ , 8 13 P' ~ I v v v v._ v ~' li:; v v. J ~ ,~ / ~ 21 22 23 24 25 26 27 29 32 35 Pyrogram Peak Figure 5-Center values for the three sagebrush palatability FCV classes determined using supervised classification. Numbers above the bars indicate the 20 most important features according to CART. 330 12 that HCA was capable of discriminating all nine locations; however, no apparent correlation to ploidy was found. Because exploratory data analysis did not reveal any relationship with ploidy, supervised modeling was used on the data set to obtain chemical models corresponding to the four euploids. A weighted feature was added to the data set which corresponded to the four euploids. The results from HCA on the supervised data set (figs. 10 and 11) show that HCA correctly classified the shadscale samples into one of four classes corresponding to 2x, 4x, 8x and lOx. The dendrogram also indicated similarities and dissimilarities between the groups of shadscale pyrograms, with 4x and 8x samples the most similar, and the 2x the most dissimilar. MVSP principal component analysis was applied to the shadscale data set in table 2. Figure 12 shows a plot of the first and second principal components. Results corroborated previous results obtained from HCA. Shadscale The shadscale samples used for this study are shown in table 2. The resulting pyrograms from the Py-GC analyses of these samples, consisting of approximately 20 peaks, were compiled into a 48 x 28 data matrix. Representative pyrograms for each different ploidy level are shown in figures 6 and 7. HCA was applied to the resulting data matrix using two different clustering methods: (UPGMA) and (CLINK). The resulting dendrograms are shown in figures 8 and 9, respectively. Except for the distance values at which the samples formed clusters, the order in which samples merged to form the dendrograms is essentially the same for both clustering methods. Because UPGMA and CLINK are based on different philosophies, the resulting clusters can be considered as well defined and not artifacts of the clustering method. Figures 8 and 9, show 8x 2x lOx 4x Figure &-Representative pyrograms from Figure 7-Representative pyrograms from Py-GC analysis of shadscale chromosome races 2x and 4x. Py-GC analysis of shadscale chromosome races ax and 1Ox. 331 Ohsia11arity Value Ohsia11arity Value 10 IS 20 10 25 15 20 25 l~ry,UT87 21 2 ~~ock f Springs, IIY 1---------, 3 ~~88 31 3 32 3 3 3 37 3 4 3 41 42 4 4 4 4 47 4 Figure &-Dendrogram from unsupervised hierarchical cluster analysis of shadscale using average linkage (UPGMA) between groups and Euclidean distance measure. Figure 10-Dendrogram from supervised hierarchical cluster analysis of shadscale using average linkage (UPGMA) between groups and Euclidean distance measure. D1ssia11arity Yalu1 10 15 Ohsia11arity Value 20 25 10 15 1 , 87 21 r : h r yUT 2 ~~ock Springs, IIY ~7 1--------~ 3 ~~ Ellery, Uf 88 31 3 32 3 3 3 37 3 4 3 41 42 4 4 4 4 47 4 Figure 9-Dendrogram from unsupervised hierarchical cluster analysis of shadscale using complete linkage (CLINK) between groups and Euclidean distance measure. Figure 11-Dendrogram from supervised hierarchical cluster analysis of shadscale using complete linkage (CLINK) between groups and Euclidean distance measure. 332 20 25 0.025 0 ~ 0.015 The FCV algorithm was used to determine the chemical features that differentiate the four classes found by HCA and PCA. Figure 13 plots the contribution of each class center (average values of each class) to each of the 28 Euclidean dimensions in the data set. Several peaks have high discriminating power among the four shadscale euploids: peaks 16 and 20 for discriminating lOx, peak 24 for discriminating 2x, peak 9 for discriminating 8x, and the absence of any peaks for discriminating 4x from the other three. 6. 2x 0 0 0 4x Sx lOx '--- ~ N 'E Cl) c 0 a. 0.005 E 0 (.) asa. '(j @ 6 ~ -0.005 .5 ~ 0:. g -0.015 § DISCUSSION 0 Big Sagebrush A feature may be important to the class structure in two ways. It may show a high variation within a class, thus high "modeling power" for that particular class, or it may be a good discriminator among classes. In unsupervised clustering, one class is split so that the variations within the new classes are minimized relative to the global variation. A feature with global modeling -0.025 -0.1 0.0 0.1 0.2 Principal Comoonent 1 Figure 12-First and second supervised principal components analysis of shadscale samples. ~ .18 2x ~ 7 10 m Bx 4x • lOx 19 22 .16 .14 s..... Q) ,.._;l f=l Q) u en en cd r--i .12 .1 u 0 ~ Q) 0 ~ cd ,.._;l en ·~ ~ 1 4 13 16 Py rogran 1 Peaks Figure 13-Center values of the four chromosome races (2x, 4x, ax, 10x) FCV classes determined using supervised classification. 333 25 2A power should, therefore, be a good discriminator at the subset or class level. Consequently, features with high modeling power according to this model should be good discriminators for taxonomy classes but not for palatability classes. The class centers from FCV, shown in figure 3, illustrate the chemical differences among the three palatability classes. Eight features of possible importance to the palatability can be identified visually as shown in figure 3 (features with a stepwise increase or decrease with palatability). The combined use of FCV and CART, shown in figure 5, resulted in a subset of 20 peaks from figure 3 ranked as important for classification (features ; : : 4 7 percent in table 4). Numbers above the bars indicate CART's ranking. The eight peaks with a stepwise increase or decrease with palatability are among the 20 peaks in the subset, with peak 20 being the most important if the samples are to be classified according to palatability. In a related study, Welch and McArthur (1986) have shown that coumarin compounds are good taxonomic indicators, as well as palatability indicators for mule deer. Further studies on peak identification should begin with a comparative study of known taxonomic and palatability indicators (such as coumarin compounds) and peak 20. This result demonstrates that Py-GC-PR is a viable method for the determination of big sagebrush palatability. its relationship to the abundance and species of insects on shadscale. A survey ofinsects associated with native shrubs during 1986 to 1989 has included collections from areas where the ploidy of shadscale is known. It should be possible to determine if insects and ploidy are correlated. If insects are shown to be to be associated (directly of indirectly) with shrub dieoff, and with ploidy, understanding the causes of dieoff would be increased. Similar correlations could be calculated when quantitative, biological data are available about plant diseases, edaphic factors, and range management (grazing, etc.). It may well be that some of these associations will be shown to be random. However, if these associations are real by using shadscale as a test case, the principles would have widespread use. For example, as guidelines for plant materials centers, production and commercialization of native seeds, range management, gathering information about poisonous range plants, and a host of other applications. CONCLUSIONS Big Sagebrush Hierarchical application of FCV discriminated among the 10 accessions. However, neither model resulting from unsupervised data analysis correlated with palatability classes. The two programs used for supervised classification employed different approaches to evaluate the chemical features. The FCV class centers identified eight features that increased or decreased stepwise with increasing palatability. When these results were combined with the feature evaluation performed in CART, one chemical feature (peak 20) proved to discriminate best for palatability; further studies on peak identification should begin with this feature. Of the three pattern-recognition programs, the most definitive information was obtained by combining FCV and CART. Given an adequate training set, the combined application of these algorithms is recommended for interpretation of complex data sets such as those resulting from Py-GC analysis of big sagebrush. Shadscale The three multivariate pattern-recognition programs (HCA, PCA, and FCV) applied to the pyrolysis-gas chromatographic data correctly classified the shadscale samples according to location, with the supervised approach showing each of the ploidy levels. Output of the FCV algorithm was used to determine pyrogram peaks responsible for discriminating among the four ploidy levels. Five features (pyrogram peaks 6, 9, 16, 20, and 24) or their absence were important in discriminating each of the ploidy levels of shadscale. The biological questions that gave rise to these chemical tests of ploidy were: Knowing that fingerprinting techniques similar to those described here have previously discriminated among range grasses with different susceptibilities to insect feeding (Windig and others 1983), can ploidies of shadscale (2x, 4x, etc.) be classified using similar methods? Can ploidy be identified all seasons of the year? Is ploidy related to the kinds and abundance of insects found in nativ~ rangelands? Are insects directly or indirectly related to dieoff of native shrubs? Results presented here answer some of these questions. Yes, shadscale ploidy can be classified using Py-GC. Discrimination of plant location was also determined, a very important observation that may assist in identifying plants that should or should not be grown in certain areas because of their adaptation characteristics. Data now on hand and being analyzed may provide answers to some of the other questions about ploidy and Shadscale The results of this study using a limited data set from four ploidy levels of shadscale (2x, 4x, 8x, and lOx) demonstrated that Py-GC-PR is capable of discerning minute biochemical differences among morphologically similar accessions of shadscale. In addition, this study demonstrates that Py-GC-PR, given a large enough training set, could classify and differentiate unknown samples of shadscale according to their ploidy levels. Determination of the chemical identity of the discriminating pyrogram peaks allows development of rapid screening methods for shadscale identification through the use of Py-GC-PR. 334 ACKNOWLEDGMENTS McArthur, E. D.; Welch, B. L. 1982. Growth rate differences among big sagebrush (Artemisia tridentata) accessions and subspecies. Journal of Range Management. 35: 396-401. Meglen, R. R. 1988. Chemometrics: its role in chemistry and measurement sciences. Chemometrics and Intelligent Laboratory Systems. 3: 17-29. Romesburg, H. C. 1984. Cluster analysis for researchers. Belmont, CA: Lifetime Learning Publications. Soderstrom, B.; Frisvad, J. C. 1984. Separation of closely related asymmetric penicillia by pyrolysis gas chromatography and mycotoxin production. Mycologia. 76: 408-419. Sharaf, M. A.; Tilman, D. L.; Kowalski, B. R. 1986. In: Elving, P. J.; Winefordner, J.D., eds. Chemometrics Vol. 82 in chemical analysis. New York: John Wiley and Sons: Chapter 6. Stutz, H. C.; Sanderson, S. C. 1983. Evolutionary studies of Atriplex: chromosome races of A. confertifolia (shadscale). American Journal of Botany. 70(10): 1536-1547. Tabachnick, B. G.; Fidell, L. S. 1983. In: Using multivariate statistics. New York: Harper and Row. Torell, J.; Evans, J.; Valcarce, R.; Smith, G. G. 1989. Chemical characterization of leafy spurge (Euplwrbia esula L.) by Curie-point pyrolysis-gas chromatographypattern recognition. Journal of Analytical Applied Pyrolysis. 14: 223-236. Valcarce, R.; Smith, G. G. 1989a. Chemical characterization of honey bees by Curie-point pyrolysis-gas chromatography-pattern recognition. Chemometrics and Intelligent Laboratory Systems. 6: 157-166. Valcarce, R.; Smith, G. G. 1989b. Pattern recognition studies of Curie-point pyrolysis-gas chromatographic data from materials important to agriculture. Journal of Analytical Applied Pyrolysis. 15: 357-372. Vogt, N. B.; Bye, E.; Thrane, K. E.; Jacobsen, T.; Benestad, C. 1989. Composition activity relationshipsCARE: Part 1. Exploratory multivariate analysis of elements, polycyclic aromatic hydrocarbons and mutagenicity in air samples. Chemometrics and Intelligent Laboratory Systems. 6:31-47. Welch, B. L.; McArthur, E. D. 1986. Wintering mule deer preference for 21 accessions of big sagebrush. Great Basin Naturalist. 46: 281-286. Welch, B. L.; McArthur, E. D.; Rodriguez, R. L. 1987. Variation in utilization of big sagebrush accessions by wintering sheep. Journal of Range Management. 40: 113-115. Windig, W.; Meuzlaar, H. L. C.; Haws, B. A.; Campbell, C. F.; Asay, K. H. 1983. Biochemical differences observed in pyrolysis mass spectra of range grasses with different resistance to Labops hesperus Uhler attack. Journal of Analytical and Applied Pyrolysis. 5: 183-198. Wold, S.; Albano, C.; Dunn, W. J., II; Edlund, U.; Esbensen, K.; Geladi, P.; Hellberg, S.; Johansson, W.; Lindberg, W.; Sjostrom, M. 1984. In: Kowalski, B. R., ed. Chemometrics: mathematics and statistics in chemistry. NATO ASI Series: NATO Science Affairs Division. New York: Reidel Publishing. The authors wish to thank R. W. Gunderson for the use of his programs FCVPC-87 and LINK; T. Jacobsen, on leave from the Brewing Industry Research Institute, Oslo, Norway, for help and useful discussions during data processing; and J. Robinson for help with the Py-GC analyses. Portions of this study were funded by the Utah Agricultural Experiment Station, Logan, UT; the Biotechnology Center, Logan, UT; Utah State University; and the USDA Forest Service, Intermountain Research Station. Journal paper No. 3885 of the Utah Agricultural Experiment Station. REFERENCES Behan, B.; Welch, B. L. 1986. Winter nutritive content of black sagebrush (Artemisia nova) grown in a uniform garden. Great Basin Naturalist. 46: 161-165. Breiman, L.; Friedman, J. H.; Olshen, R. A.; Stone, C. J. 1984. In: Bickel, P. J., ed. Classification and regression trees. Belmont CA: Wadsworth International: Chapter 8. Duewer, D. L.; Kowalski, B. R.; Fasching, J. L. 1976. Improving the reliability of factor analysis of chemical data by utilizing the measured analytical uncertainty. Analytical Chemistry. 48: 2002. Dunn, G.; Everitt, B.S. 1982. An introduction to mathematical taxonomy. New York: Cambridge University Press. Gunderson, R. W. 1984. FCV-manual. Logan, UT: Utah State University, Dept. of Electrical Engineering. Gunderson, R. W.; Thrane, K.; Nilson, R. D. 1988. A falsecolor technique for display and analysis ofmultivariable chemometric data. Chemometrics and Intelligent Laboratory Systems. 3: 119-131. Irwin, W. J. 1982. In: Analytical pyrolysis: a comprehensive guide. Chromatographic Science Series Vol. 22. New York: Marcel Dekker. Jacobsen, T.; Gunderson, R. W. 1987. In: Piggot, J. J., ed. Statistical procedures in food research. Elsevier: Chapter 10. Jerman-Blazic, B.; Fabic-Petrac, I.; Randic, M. 1989. Evaluation of the molecular similarity and property prediction for QSAR purposes. Chemometrics and Intelligent Laboratory Systems. 6: 49-63. Jurs, P. C. 1986. Pattern recognition used to investigate multivariate data in analytical-chemistry. Science. 232: 1219-1224. Knudson, E. A.; Duewer, D. L.; Christian, G. D.; Larson, T. V.. 1977. In: Kowalski, B. R., ed. Chemometrics: theory and application. Washington, DC: ACS Symposium Series 52. McArthur, E. D.; Plummer, A. P. 1978. Biogeography and management of native western shrubs: a case study, Section Tridentatae of Artemisia. Great Basin Naturalist Memoirs. 2: 229-243. McArthur, E. D.; Pope, C. L.; Freeman, D. C. 1981. Chromosomal studies of subgenus Tridentatae of Artemisia: evidence for autoploidy. American Journal of Botany. 68: 589-605. 335