– Small Molecule Drug Discovery Binding by NMR BCMB/CHEM 8190
advertisement

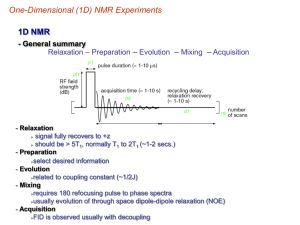
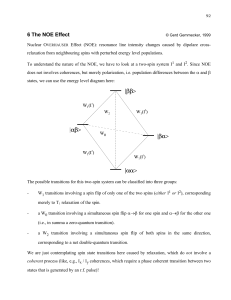
Drug Discovery – Small Molecule Binding by NMR BCMB/CHEM 8190 SAR by NMR Shuker, Hajduk, Meadows, Fesik, Science, 274, 1531 (1996) KA KB GAB = GA + GB, KAB RTln(K) = - GAB , KAB = KAx KB KA = 2 x 103, KB = 5 x 103 , KAB = 1 x 107 Chemokine CCL5 Interacting with Chondroitin Sulfate Oligomer – Where is the Oligomer? Crystal Structure: Murooka et al, JBC 281:25184 (2006) 110.0 105.0 Titration of CCL5 with Chondroitin Sulfate Pentamer 15N PPM 115.0 120.0 N46 K45 T43 R47 I24 V49 125.0 V42 Q48 130.0 R47 W57 10.5 10.0 9.5 9.0 8.5 8.0 1H PPM 7.5 7.0 6.5 6.0 Dissociation constant & Stoichiometry Determined P/L=1:1 P/L=2:1 Langmuir isotherm is often mistakenly used: correct binding formula: FB [ P] [Tb3 ] K D ([P] [Tb 3 ] K D ) 2 4[ P][Tb3 ] 2[ P] Docking Structures Using HADDOCK De Vries, …, Bonvin, A. (2007) Prot.-Struct. Funct. and Bioinfo. 69, 726-733. SAR by NMR Examples From Stockman, (1998) Progress in NMR Spec, 33, 109-151 FKBP Stromelysin Transferred NOEs in Drug Discovery • Determine the geometry of a bound ligand (Sayers and Prestegard, Biophysical J, 81, 0000 (2001)) • Screening of a library of potential ligands (Meinecke and Meyer, J. Med. Chem, 44, 3059 (2001)) • NMR as a tool for structure-based drug design (B. Stockman, Prog. NMR Spec., 33, 109 (1998)) Transfer NOE sipb sijf K-1 sijb K1 Protein sjpb Ligand *Cross-relaxation rate c, r, 0 *Chemical exchange fast w.r.t cross-relaxation rate and chemical shift scale *Observed NOE is weighted average of free and bound states *Change in sign of NOE upon change in molecular weight *Spin Diffusion and on, off rates can complicate interpretation NOEs from large and small molecules have opposite signs 0.5 0.0 -1.0 c = 1.1(1/0) Rates of transfer (s) are proportional to c for large molecules. Observe: NOE(obs) = p(bound)•s(bound) + p(free)•s(free) NOE from bound state dominates for p(bound)/p(free) > 0.05 -Me Mannose Bound to MBP Transfer NOE studies of Trimannoside with MBP Man II f 4’ 1 2 y Man III 3’ Man I Methyl 3,6-di-O-(-D-mannopyranosyl)-(-D-mannopyranoside) Schematic representation of trimannoside core structure depicting some of the intra and inter-residue NOEs observed Pulse Sequence for Selective 1D NOE 90x 180y 180y 90x 90+/-x Mix Gradient z x z y x y x a b z a b y x z z y x y Behavior for selected resonance from two different volume elements on +x 90° pulse. Vectors return to +z with –x 90° pulse. c Tris 0.1 mM MBP, 1 mM Trimannoside trNOE b NOE -OCH3 Selective saturation H3’ H1 H1’ H1’’ H2 a H4’ Selective saturation NOE difference spectra for trimannoside STD References • Saturation Transfer Difference, Mayer, M & Meyer, B. JACS, 123, 6108-6117 (2001) • Perspectives on NMR in drug discovery: a technique comes of age, Pellecchia M; et al., Nature Reviews Drug Discovery, 7: 738-745 (2008) • STD-NMR: application to transient interactions between biomolecules-a quantitative approach, Angulo J and Nieto PM, European Biophysics J., 40:1357-1369 (2011) • NMR Screening and Hit Validation in Fragment Based Drug Discovery, Campos-Olivas R. Current Topics Med. Chem., 11:43-67 (2011) Spin Diffusion is Efficient in Macromolecules – Saturation of Protein Spins transfers to Ligands 1/T1,2 = ij Ji (i) | Dij|2 , Ji (i) = 2c/(i2 c2 + 1) 1/T2 (const) Ji (i) | Di(I+I-, I-I+) |2 for large c H1 H1 H1 H1 H1 H1 H1 H1 H1 Saturation Transfer Difference Method (STD) for Ligand Screening Peptides Used in STD Experiments STD Results STD can also be used in screening






![Job Evaluation [Opens in New Window]](http://s2.studylib.net/store/data/009982944_1-4058a11a055fef377b4f45492644a05d-300x300.png)