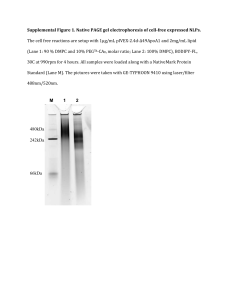
Alignment Media Protocol AIK, updated 4/8/2004: 1. DMPC/DHPC ester linkage bicelles:
advertisement

Alignment Media Protocol AIK, updated 4/8/2004: 1. DMPC/DHPC ester linkage bicelles: 2. DMPC/DHPC ether bicelles (hydrolysis resistant): 3. PEG a. C12E5 b. C8E5 4. CPyBr 5. Gels DMPC/DHPC ester linkage bicelles: q=3:1 25% w/v lipid 25 g/100mL solvent. To make 1mL bicelles, need 250mg lipid. DMPC = 687g/mol DHPC =453.3g/mol DMPC 3*687 = 2061 DHPC 1*453.3=453.3 2062/453.3=4.55 4.55+1=5.55 0.25/5.55=0.0450721 mol fraction 0.045*4.55 = 205 mg DMPC 0.045*1 = 45 mg DHPC Mix each lipid separately in 500uL buffer. Make sure buffer contains D2O!! Vortex thoroughly. DHPC dissolves quickly, DMPC is insoluble – get a white, chalky suspension. When both solutions are homogeneous, add DHPC to DMPC and vortex vigorously. Small, white chunks may form. Freeze with liquid N2 or acetone/dry ice mixture (regular ice/water bath not cold enough to rapidly freeze bicelles). Thaw to RT, vortex. Sonicate. Repeat at least 3 times and continue process until sample is clear at RT, no chunks. If chunks remain, place bicelles in –80ºC freezer 1-2 hours, thaw, vortex. If chunks still pleasant, try to siphon only clear, dissolved materials for bicelle use. DMPC/DHPC ether bicelles (hydrolysis resistant): q=3:1 15% w/v lipid 15 g/100mL solvent. To make 1mL bicelles, need 150 mg lipid. DMPC = 649.97 g/mol DHPC = 425.54 g/mol 3*649.97 = 1949.91 1*425.54 = 425.54 1949.91/425.54 = 4.58 4.58+1 = 5.58 0.15/5.58 = 0.02687, mole fraction 0.02687*4.58 = 123 mg DMPC 0.02687*1 = 26.9 mg DHPC 15 g/100mL = 90 mg/ 600 L Want to make only 600 L of 15% bicelles 123*60% = 62.2 mg DMPC 26.8*60% = 12.7 mg DHPC (This protocol was used for a real sample at pH 5.0. Aligned from 25-30c, isotropic above and below this temperature range). Mix each lipid separately in 300 L buffer – ensure buffer contains D2O!!!. Vortex thoroughly. DHPC should dissolve easily. DMPC will NOT dissolve but rather forms milky-white suspension. When both samples are homogeneous, add DHPC to DMPC, vortex well. Small, white chunks may form. Freeze with liquid N2 or acetone/dry ice mixture (regular ice/water bath not cold enough to rapidly freeze bicelles). Thaw to RT, vortex. Sonicate. Repeat at least 3 times and continue process until sample is clear at RT, no chunks. If chunks remain, place bicelles in –80ºC freezer 1-2 hours, thaw, vortex. If chunks still pleasant, try to siphon only clear, dissolved materials for bicelle use. DHPC: hygroscopic. Pump dry in lyophilizer before use. Alternatively, pipet some that is stored in chloroform/methanol, remove w/ N2 and pump dry in vacuum pump (do not use lyophilizer for organic solvents). Always allow powder samples to come to room temp before measuring. Vnmr (common to all LCs, may not need to adjust temp at all, may need to find temp and temp range of alignment) Check 2H alignment in probe. Lock, shim at 25c. Turn off lock power, recable for 2H observe through lock channel. Make sure tn (transmitter nucleus) is set to 2H, use tpwr ~45 dB and pw ~10 us. The pw is actually MUCH longer than this, but we’re not trying to be quantitative: we want alignment. Collect spectrum and svf. Settemp(35). Software doesn’t know which sample to lock onto now b/c there are two lock frequencies. Don’t bother locking and shimming unless you have a lot of time: Bo inhomogeneity shouldn’t prevent you from seeing if your sample is aligned or not. Collect spectrum. To monitor your sample as temp increases (and a fun way to kill time), array the parameter nt for about 100 steps of 1 scan each. You should see the phase transition. Look for 2H quadrupolar splittings. Measure the splittings. If you want to check the temp range of alignment, set vttype=2 and array temp as you would array any other parameter. Also array pad to give the sample time to equilibrate btwn each temp. Set up the temp array and the pad array separately. Then type array=’(pad,temp)’ and the two parameters will be arrayed simultaneously. C12E5 (pentaethylene glycol monododecyl ether), Ruckert and Otting, JACS 122:7793 (2000). Available from Sigma. 4.25% w/w (includes mass of solvent, lipid, but NOT hexanol) r = 0.87 (r is molar ratio C12E5:hexanol) stable over wide pH range Mix in this order: 25uL C12E5, 500uL water or buffer (D2O included), vortex very well. C12E5 is viscous – do not pull the pipette out of the bottle immediately after filling pipette tip. Wait at least 15sec to make sure C12E5 gets into pipette tip. Add 9uL hexanol in 3uL or 4.5uL increments, vortexing well after each addition. Solution goes from clear to milky, turbid. Then to translucent and viscous w/ lots of bubbles (upon vortexing). When liquid crystalline, solution looks blue-ish in color. This phase does align in the magnet but could be biphasic, heterogeneous. If solution becomes cloudy, milky again and very fluid, you have added too much hexanol and have passed nematic phase. Careful pipetting technique is essential! 2 H splitting at 25C should be about 23-26 Hz but may change upon addition of biomolecule to align. The higher the concentration of “lipid” you use, the more hexanol you may have to use. You will have to experimentally determine the correction ration of C12E5:hexanol. C8E5 (Pentaethylene glycol octylether) 5% w/w (includes mass of solvent, lipid, NOT octanol) r = 0.84 (r = molar ratio C8E5:octanol) (C8E5 more soluble in water than C12E5) e.g. 5% C8E5 w/ r=0.84 aligns at 25C to 24-26Hz 2H quad splitting Add 176 uL C8E5 to Eppendorf tube. Add 640 uL buffer (this buffer does not have D2O), 100 uL D2O, 64 uL octanol in 8 uL increments. Another C8E5 protocol that also works: 51uL C8E5, 16uL octanol, 100uL D2O (or if D2O already in buffer ignore this step and add 100uL to buffer amount), 825 uL buffer. 4.83w/w% C8E5, r=1.35. 2H splits to 32Hz. CPBr (cetylpyridinium bromide) Barrientos et al, JBNMR 16:329-337 (2000). 6.5% (w/v) Cetylpyridinium bromide 25 mM NaBr hexanol r, CPBr:hexanol, = 1:1.33 (w/w) Stable over wide pH range