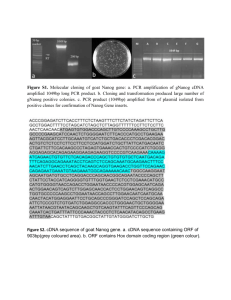
The Homeoprotein Nanog Is Required for Maintenance
advertisement

Cell, Vol. 113, 631–642, May 30, 2003, Copyright 2003 by Cell Press The Homeoprotein Nanog Is Required for Maintenance of Pluripotency in Mouse Epiblast and ES Cells Kaoru Mitsui,1 Yoshimi Tokuzawa,1 Hiroaki Itoh,1 Kohichi Segawa,1 Mirei Murakami,1 Kazutoshi Takahashi,1 Masayoshi Maruyama,1 Mitsuyo Maeda,2 and Shinya Yamanaka1,* 1 Laboratory of Animal Molecular Technology Research and Education Center for Genetic Information Nara Institute of Science and Technology Nara 630-0192 Japan 2 First Department of Anatomy Osaka City University Medical School Osaka 545-8585 Japan Summary Embryonic stem (ES) cells derived from the inner cell mass (ICM) of blastocysts grow infinitely while maintaining pluripotency. Leukemia inhibitory factor (LIF) can maintain self-renewal of mouse ES cells through activation of Stat3. However, LIF/Stat3 is dispensable for maintenance of ICM and human ES cells, suggesting that the pathway is not fundamental for pluripotency. In search of a critical factor(s) that underlies pluripotency in both ICM and ES cells, we performed in silico differential display and identified several genes specifically expressed in mouse ES cells and preimplantation embryos. We found that one of them, encoding the homeoprotein Nanog, was capable of maintaining ES cell self-renewal independently of LIF/Stat3. nanog-deficient ICM failed to generate epiblast and only produced parietal endoderm-like cells. nanog-deficient ES cells lost pluripotency and differentiated into extraembryonic endoderm lineage. These data demonstrate that Nanog is a critical factor underlying pluripotency in both ICM and ES cells. Introduction Embryonic stem (ES) cells derived from inner cell mass of mammalian blastocysts grow rapidly and infinitely while maintaining pluripotency, the ability to differentiate into all types of cells (Evans and Kaufman, 1981; Martin, 1981). These properties of ES cells are maintained by symmetrical self-renewal, producing two identical stem cell daughters upon cell division (Burdon et al., 2002). These properties have raised the hope that ES cells could be used to treat a host of degenerative diseases such as Parkinson’s disease and diabetes (Smith, 1998). This hope was accelerated by the generation of pluripotent cells from human blastocysts (Thomson et al., 1998). However, human ES cells also raised substantial ethical issues since human embryos have to be destroyed to generate ES cells (Colman and Burley, 2001; Lachmann, 2001; Mieth, 2000). One solution to *Correspondence: shinyay@gtc.aist-nara.ac.jp avoid such ethical issues is to generate pluripotent cells directly from somatic stem and other cells. The first step toward this goal is to understand molecular mechanisms underlying pluripotency. Leukemia inhibitory factor (LIF) has been utilized to maintain the symmetrical self-renewal of mouse ES cells (Smith et al., 1988; Williams et al., 1988). LIF causes heterodimer formation of the LIF receptor and gp130, which resulted in activation of the tyrosine kinase Jak (Ernst et al., 1996). The activated Jak phosphorylates several tyrosines of gp130, which serve as docking sites for proteins containing Src homology 2 (SH2) domains, including the signal transducer and activator of transcription (Stat) family of transcription factors (Ihle, 1996). Multiple lines of evidence indicated that Stat3 is the key downstream transcription factor of the LIF/gp130 pathway in ES cells. Forced expression of a dominantnegative Stat3 mutant caused differentiation of ES cells even with the presence of LIF (Niwa et al., 1998). Point mutation of the tyrosine residue of gp130 responsible for the Stat binding abrogated the ability of LIF to maintain self-renewal, while other tyrosines were dispensable (Ernst et al., 1999; Matsuda et al., 1999). ES cells expressing a fusion molecule consisting of Stat3 and estrogen receptor could be maintained pluripotent by the estrogen derivative tamoxifen (Matsuda et al., 1999). Gene targeting experiments also demonstrated the importance of Stat3 for ES cell self-renewal (Raz et al., 1999). However, mutant embryos deficient in the LIF/gp130/ Stat3 pathway apparently form normal ICM; LIF-deficient mice develop normally (Stewart et al., 1992), while LIF receptor-deficient mice showed perinatal lethality (Li et al., 1995; Ware et al., 1995). Embryos deficient in gp130 progressively die between 12.5 days postcoitum (dpc) and term (Yoshida et al., 1996). Stat3-deficient embryos developed into the egg cylinder stage until embryonic day (E)6.0 and showed a rapid degeneration between E6.5–E7.5 (Takeda et al., 1997). A role of LIF/ gp130 in preimplantation embryos is evident only for prolonged survival of blastocysts during diapause (Nichols et al., 2001). Furthermore, LIF is not required for selfrenewal of human ES cells (Reubinoff et al., 2000) and several mouse ES cell lines (Berger and Sturm, 1997; Dani et al., 1998). In addition, Stat3 is expressed in wide ranges of cell types and, in some cases, is essential for differentiation (Nakajima et al., 1996). These results argue that LIF/gp130/Stat3 are not fundamental for pluripotency and predict the existence of a novel pathway(s) that maintains pluripotency in both ICM and ES cells. Several transcription factors have been shown essential for pluripotency in ICM, but none of them function independently of LIF/Stat3. Oct3/4 is a POU family transcription factor specifically expressed in ES cells, preimplantation embryos, epiblast, and germ cells (Okamoto et al., 1990; Scholer et al., 1990). Inactivation of oct3/4 in embryos and ES cells resulted in loss of pluripotency and spontaneous differentiation into trophoblast lineage (Niwa et al., 2000). However, ES cells expressing Oct3/4 constitutively from an exogenous promoter still required Cell 632 Table 1. Top 20 Genes with Highest Enrichment in ES Cells Identified by Digital Differential Display Unigene Frequency ES/EC cell Frequency Others Symbol Mm.139314 Mm.10205 Mm.5090 Mm.157658 Mm.46472 Mm.5180 Mm.6047 Mm.28369 Mm.17031 Mm.3396 Mm.45676 Mm.249524 Mm.23310 Mm.18154 Mm.13433 Mm.47904 Mm.4213 Mm.913 Mm.28388 Mm.158190 0.00295 0.0028 0.00079 0.00076 0.00076 0.00073 0.00067 0.00049 0.0004 0.00033 0.00033 0.00033 0.0003 0.00027 0.00027 0.00027 0.00024 0.00024 0.00024 0.00021 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ecat2 utf1 cripto ecat1 hnRNB-G Nr0b1/dax-1 ecat4/nanog ecat3/fbx15 oct3/4 Zfp42/rex1 EST ecat5/ERas zfp296 tcl1 ecat7 ecat8 ecat9 brachyury (T ) tex20 ecat6 LIF for self-renewal (Niwa et al., 2000). Overexpression of oct3/4 induced differentiation similar to that caused by stat3 inactivation, indicating crosstalk between the two transcription factors. Two other transcription factors, Sox2 (Avilion et al., 2003) and FoxD3 (Hanna et al., 2002), were recently shown essential for pluripotency in embryos. Since both Sox2 and FoxD3 cooperatively function with Oct3/4 (Guo et al., 2002; Yuan et al., 1995), it is likely that they also have crosstalk with Stat3. The purpose of this study is to identify the LIF/Stat3independent factor(s) that underlies pluripotency in both ICM and ES cells. To this end, we conducted digital differential display to identify genes expressed in ES cells as specifically as oct3/4. We explored functions of these genes by constitutive expression in ES cell and targeted gene disruption. Here, we report that one of them, which we now call Nanog for Tir Na Nog (land of the ever young), maintains pluripotency of both ICM and ES cells independently of LIF/Stat3. Results Identification of ecat by Digital Differential Display To identify candidates of the LIF/Stat3-independent factor(s) essential for pluripotent cells, we performed digital differential display (http://www.ncbi.nlm.nih.gov/UniGene/ info_ddd.shtml) to compare expressed sequence tag (EST) libraries from mouse ES cells and those from various somatic tissues. A number of genes were found overrepresented in ES cell-derived libraries (Table 1). Among twenty genes with highest enrichment were oct3/4, utf1 (Okuda et al., 1998), and rex1 (Rogers et al., 1991), which had been experimentally identified as ES cell-specific markers. Northern blot analyses confirmed ES cell-specific expression in nine uncharacterized genes (Figure 1). We designated these genes ecat, for ES cell associated transcripts. Figure 1. Northern Blot Analyses of ecat Genes Five micrograms of total RNA was analyzed with mouse cDNAs of each ecat and H-ras as probes. Differentiation of ES cells was induced with retinoic acid (3 ⫻ 107 M) for 5 days. LIF-Independent Self-Renewal by Ecat4/Nanog We expressed ecat, oct3/4, or sox2 constitutively in ES cells to explore their ability to render ES cells independent of LIF for self-renewal. To this end, we utilized the supertransfection system, in which more than 90% of transfected cells expressed the gene of interest, allowing functional analyses of polyclonal population (Aubert et al., 2002; Niwa et al., 2002). As has been reported, forced expression of Oct3/4 resulted in differentiation of ES cells (data not shown). Ecat9 and Sox2 induced massive cell death. We were able to establish cells constitutively expressing the other eight Ecats. When cultured with LIF, all of them showed normal morphology. When cultured without LIF, all but one differentiated normally as judged by flattened morphology (Figure 2A) and reduced oct3/4 expression (Figure 2B). In contrast, cells constitutively expressing ecat4 did not show such a morphological change even after prolonged culture (⬎1 month) without LIF (Figure 2A). Expression of oct3/4 also remained normal (Figure 2B). Because of this unique property, we renamed ecat into nanog from Tir Na Nog (land of the ever young). These (CAG-nanog) cells expressed significantly more nanog transcripts (Figure 2B) and proteins (Figure 2C) than control cells. The nanog expression sustained even after prolonged culture without LIF. Stat3 activation kinetics was unchanged (Figure 2D). Growth speed was faster than control cells but significantly decreased by LIF removal (Figure 2E). These data demonstrated that constitutive expression of Nanog maintains the undifferentiated state of ES cells independently of the LIF/Stat3 pathway. Stat3-Independent Pluripotency Maintenance Factor 633 Figure 2. Forced Expression of Nanog from a Constitutive Promoter (A) Morphology of MG1.19 cells transfected with the parent plasmid (mock) or the Nanog expression vector (CAG-nanog). Cells were grown with or without LIF for three passages after puromycin selection. (B) Expression levels of nanog, oct3/4, and nat1 in cells described in (A) were determined by RT-PCR. (C) Lysates were isolated from cells described in (A) and blotted with ␣-Nanog antiserum or ␣-Cdk4 antibody (Santa Cruz, sc-260-G). Lower bands detected by ␣-Nanog antiserum are likely originated from proteolytic products. (D) LIF was removed for 60 min and was added back to culture medium. Cell lysates were collected at the indicated time points and blotted with ␣-Nanog antiserum, ␣-Oct3/4 antibody (Santa Cruz, sc-5279), ␣-phospho-Stat3 (Tyr705) antibody (Cell Signaling Technology, #9131), and ␣-Stat3 antibody (Santa Cruz, sc-482). (E) Cells (10,000 cells per well of 24-well plates) were cultured with or without LIF for 5 days and cell numbers were counted. Shown are averages and standard deviations of four independent experiments. A Unique Homeoprotein Encoded by nanog The nanog cDNA consisted of 2184 nucleotides (nt) and contained a single open reading frame encoding polypeptide of 305 amino acids. It has a long 3⬘ untranslated region of 1077 nt containing a B2 repetitive element (data not shown). Homology search revealed that the predicted Nanog protein contains a homeobox domain (Figure 3A). Phylogenic analysis showed that the Nanog Cell 634 Figure 3. Sequence and Expression of Nanog (A) Amino acid sequence of mouse Nanog is compared with human hypothetical protein FLJ12581 and mouse Nkx-2.3. Homeodomains are shown by closed boxes, in which numbers indicate amino acid identity with Nanog. Also shown are Trp repeat, TN domain, and NK2-SD domain. Tyrosine shown is strictly conserved among homeodomains of the NK-2 family, but not in Nanog or FLJ12581. (B) RT-PCR analysis showing expression profiles of mouse nanog, oct3/4, and nat1. Lanes: 1, undifferentiated MG1.19 ES cells; 2, differentiated MG1.19 ES cells; 3, undifferentiated RF8 ES cells (RT minus control); 4, undifferentiated RF8 ES cells; 5, differentiated MG1.19 ES cells; 6, brain; 7, heart; 8, kidney; 9, testis; 10, spleen; 11, muscle; 12, lung; 13, stomach; 14, ovary; 15, thymus; 16, liver; and 17, skin. (C) Lysate was collected from four ES cell lines, maintained undifferentiated (indicated by U) or treated with retinoic acid for 5 days (indicated by D). Western blot was performed with ␣-Nanog antiserum or ␣-Cdk4 antibody. homeobox is most similar to those of the Nk-2 gene family (Harvey, 1996). However, the amino acid identity is less than 50%. Furthermore, Nanog has valine at the position corresponding to tyrosine strictly conserved among the NK-2 family. The TN domain and the NK-2specific domain, which are also conserved among the NK-2 family, do not exist in Nanog. These data indicate that Nanog is distinct from the NK-2 family. Amino acid identities of the Nanog homeodomain with other homeoproteins expressed in ES cells, namely Oct3/4, Pem (Fan et al., 1999), and Ehox (Jackson et al., 2002), are less than 20% (data not shown). However, Nanog is identical to a homeodomain protein recently described as being expressed selectively in ES cells (Wang et al., 2003). Nanog also contains a stretch of sequence in which TrpX-X-X is repeated ten times in tandem. The function(s) of the Trp-rich repeats is not known. Transcripts of nanog were not detectable in twelve somatic organs, including testis and ovary, by either Northern blot (Figure 1) or RT-PCR (Figure 3B). Western blot with ␣-Nanog rabbit antiserum, which we raised against histidine-tagged Nanog protein, detected a specific band of expected size (35 kDa) in four independent ES cell lines: RF8 (Meiner et al., 1996), MG1.19 (Gassmann et al., 1995), J1 (Li et al., 1992), and CGR8 (Nichols et al., 1990) (Figure 3C). Retinoic acid treatment decreased Nanog protein levels in all four lines. Recently, Wang et al. (2003) reported the identification and expression analysis of the same gene as nanog. Consistent with our data, they demonstrated specific expression in ES cells and preimplantation embryos. DNA microarrays analyses demonstrated that nanog is expressed in ES cells, but not in neural or hematopoietic stem cells (Ramalho-Santos et al., 2002). Database analyses showed that the human hypothetical protein FLJ12581 is most similar to Nanog (Figure 3A). Although overall amino acid identity is relatively low (52%), its homeodomain is 85% identical to mouse Nanog. FLJ12581 also contains Trp-rich repeats, in which Typ-X-X-X is repeated eight times. In addition, FLJ12581 contains an Alu repetitive element in the 3⬘ untranslated region. FLJ12581 is located on chromosome 12, the syntenic region of mouse chromosome 6 where nanog is located. Blast search against the human dbEST identified EST clones corresponding to FLJ12581 in libraries from NT2 human teratocarcinoma cells, germ cell and testis tumors, marrow, and other tumors. No EST clones was detected in libraries from normal somatic tissues except those corresponding to the Alu element. Northern blot analysis did not detect FLJ12581 Stat3-Independent Pluripotency Maintenance Factor 635 transcripts in 12 adult organs (data not shown). These data indicate that FLJ12581 is the human nanog ortholog. Targeted Disruption of the Mouse nanog Gene in ES Cells Analysis with mouse genome databases demonstrated that the nanog gene consisted of four exons. We generated two targeting vectors that replaced the homeodomain with either a fusion of -galactosidase and the neomycin-resistance gene (geo) (Mountford et al., 1994) or the hygromycin-resistant gene (hygr) (Figure 4A). These vectors were introduced into RF8 ES cells by electroporation, and drug-resistant colonies were screened for homologous recombination. With the geo targeting vector, 27 out of 96 colonies were found positive by PCR (data not shown). With the hygr vector, 8 out 27 were positive. Homologous recombination was confirmed with Southern blot (Figure 4B) and PCR (Figure 4C). X-Gal staining of heterozygous ES cells confirmed that nanog is expressed in all undifferentiated ES cells but repressed by retinoic acid treatment (Figure 4E). To obtain homozygous mutant ES cells, we introduced the hygr vector into heterozygous clones established by the geo vector. In 34 out of 83 hyg-resistant clones, homologous recombination of the hygr vector was confirmed. Among these, 11 clones were found homozygous for nanog mutation by Southern blot (Figure 4B) and PCR (Figure 4C). Northern blot (Figure 4D) and Western blot (data not shown) analyses confirmed the absence of expression in homozygous cells. In the other 23 clones, the hygr vector replaced the geo cassette. We also introduced the geo vector into heterozygous clones established by the hygr vector. In 114 out of 397 G418-resistant clones, homologous recombination of the geo vector was confirmed. Among these, six clones were found homozygous. Cells deficient in nanog obtained by both orders showed identical phenotypes. Loss of Pluripotency and Extraembryonic Endoderm Differentiation in nanog Null Cells Cells deficient in nanog were distinct in morphology from wild type and heterozygous ES cells. When maintained on STO feeder cells, they were larger and roundshaped (Figure 5A); when cultured without feeder cells, they further flattened and showed parietal endodermlike morphology. Cells deficient in nanog grew significantly slower that heterozygous ES cells (Figure 5B) but could be serially passaged for at least 2 months (data not shown). Northern blot (Figure 5C) and RT-PCR (Figure 5D) analyses showed that nanog null cells expressed a number of endoderm transcription factors, parietal endoderm markers, and visceral endoderm markers. In contrast, mesoderm marker brachyury (t ), trophoblast marker cdx2, primitive ectoderm marker fibroblast growth factor (fgf)-5, or the neuroectodermal marker islet-1 (isl1) were not induced. Pluripotent cell markers oct3/4 and rex1 were greatly reduced though not completely eliminated when cultured on STO feeder cells. The persistence of low oct3/4 expression may be related to its proposed role in primitive endoderm specification (Palmieri et al., 1994). When cultured without feeder cells, oct3/4 and rex1 were eliminated, while gata6 was further induced (data not shown) consistent with their parietal endoderm-like morphology (Figure 5A). These data indicated that nanog disruption specified ES cell fate exclusively into extraembryonic endoderm lineages. Establishment of nanog-Deficient Mice To study in vivo function(s) of Nanog, we injected geo heterozygous ES cells into C57BL/6 blastocysts. Germline transmission was obtained from three independent ES clones. Heterozygous mutant mice were normal in gross appearance and fertile (data not shown). X-Gal staining of the preimplantation embryos detected geo expression in the inner cell mass of blastocysts and the central portion of the morula, but not in 32-cell stage or earlier embryos (Figure 6A). Lack of Epiblast in nanog Null Embryos Among 231 mice born from heterozygous intercrosses, 80 were wild-type and 151 were heterozygous. Since no homozygous offspring was obtained, we analyzed embryos of various stages. Among 36 decidual swellings obtained at 7.5 dpc, 14 contained wild-type embryos and 16 contained heterozygous mutants. Six deciduae were empty. We therefore histologically examined E5.5 embryos. Of 33 embryos examined, 26 showed normal morphology (Figure 6B). However, seven embryos appeared to consist entirely of disorganized extraembryonic tissues with no discernible epiblast or extraembryonic ectoderm. The high incidence (7 out of 33) indicates that these abnormal embryos are nanog null. They are different from foxD3 null embryos that do have epiblast at this stage (Hanna et al., 2002) and Sox2 null embryos that lack epiblast but showed expansion of extraembryonic ectoderm (Avilion et al., 2003). At 3.5 dpc, we found that 3 out of 15 blastocysts were homozygous and indistinguishable from normal embryos. When cultured on gelatin-coated plate, however, ICM of nanog null blastocysts failed to proliferate (Figure 6C). To further clarify the fate of mutant ICM, we removed trophectoderm by immunosurgery. ICM deficient in nanog did not persist as undifferentiated masses in vitro but differentiated completely into parietal endoderm-like cells within 4 days (Figure 6D). In contrast to oct3/4 null ICM (Nichols et al., 1998), no trophoblast differentiation was observed. These data demonstrated that Nanog is essential for maintenance of pluripotency of ICM at a stage after the initial requirement for Oct3/4. Identification of DNA Recognition Sequence of Nanog We determined DNA recognition sequence of Nanog by systematic evolution of ligands by exponential enrichment (SELEX). By selecting random olionucleotide with the fusion protein consisting of maltose binding protein (MBP) Nanog fusion proteins (see Experimental Procedures for details), we identified a consensus sequence (C/G)(G/A)(C/G)C(G/C)ATTAN(G/C) (Figure 7A). The highest conservation was observed with the tetramer ATTA, which is common for DNA recognition sequences of many homeobox transcription factors. This consensus sequence is different from that of the NK-2 family, T(C/T) Cell 636 Figure 4. Targeted Disruption of the Mouse nanog Gene (A) Structures of the mouse nanog gene, targeting constructs, and correctly targeted loci. Positions of the probe and ScaI recognition sites (S) used for Southern blot were shown. Arrows indicate primers for PCR analyses. (B) Southern blot with the 3⬘ probe produced an 11 kb band from the wild type locus, a 15.4 kb band from the geo-targeted locus, and a 7.3 kb band from the hygr targeted locus. (C) Specific amplification by PCR with primers 6047-insS1.2 (CACCTACCACCATGCCAGGCTGAGAATGTC) and 6047-RACE1 (AGCTGGCATCG GTTCATCATGGTACA) produced a 3.2 kb band from the wild type locus, an 8.5 kb band from the geo targeted locus, and a 4.2 kb band from the hygr targeted locus. (D) Northern blot analysis confirmed the absence of nanog transcripts in knockout cells. (E) X-Gal staining of geo/⫹ ES cells that were maintained undifferentiated on STO cells or induced to differentiate by retinoic acid (RA). AAGTG (Harvey, 1996), consistent with the amino acid differences between Nanog and NK-2. Gel-mobility shift assay showed that mobility of consensus oligonucleotides was retarded by extracts of F9 embryonic carcinoma cells (Figure 7B) and RF8 ES cells (data not shown). The shift in mobility was abolished by excess amount of unlabeled consensus oligonucleotides, but not by those with mutations in ATTA. Discussion Prior to this study, LIF/gp130/Stat3 was the only known biochemical pathway that could maintain self-renewal of mouse ES cells (Burdon et al., 2002). However, LIF/ gp130/Stat3 is dispensable for development of ICM and self-renewal of human ES cells, suggesting that this pathway is not fundamental for pluripotency (Nichols Stat3-Independent Pluripotency Maintenance Factor 637 Figure 5. Analyses of nanog Null Cells (A) Morphology of wild type RF8 cells grown on STO feeder cells (WT) and nanog null cells (KO) grown with or without STO feeder cells. (B) Three clones of heterozygous cells (⫹/⫺) and homozygous cells (⫺/⫺) were plated at 1 ⫻ 104 cells per well of 24-well plates. Cell numbers were counted after 2, 4, and 6 days. (C) Total RNA was isolated from parent RF8 cells (WT), four clones with random integration of the geo vector (⫹/⫹), four heterozygous clones (⫹/⫺), and four homozygous clones (⫺/⫺). Expression of marker genes was determined by Northern blot analyses. (D) Total RNA was isolated from wild-type RF8 cells (nanog, WT), nanog null cells (nanog, KO), ZHBTc4 cells (Niwa et al., 2000) cultured without tetracycline (oct3/4, WT), and ZHBTc4 cells cultured with tetracycline for 96 hr, which differentiated into trophoblast (oct3/4, KO). Expression of marker genes was determined by RT-PCR. et al., 2001). In this study, we demonstrated that the homeoprotein Nanog can maintain self-renewal of ES cells independently of the LIF/gp130/Stat3 pathway. Furthermore, we showed that deletion of the nanog gene resulted in loss of pluripotency in both ICM and ES cells. In the accompanying paper, Chambers et al. showed that Nanog has an ability to maintain not only undifferentiated state, but also pluripotency independently of LIF Cell 638 Figure 6. Analyses of nanog Null Embryos (A) X-Gal staining of preimplantation embryos obtained from intercross of heterozygous geo mice. (B) Decidual swellings obtained from heterozygous intercross were removed from uterus, fixed with 10% buffered formalin, and embedded in paraffin. Sections (5 m each) were stained with hematoxylin and eosin. (C) Blastocysts obtained from heterozygous intercrosses were grown on gelatin-coated plate with ES cell medium. After 10 days, morphology was recorded and genotype was determined by PCR. All homozygous blastocysts showed similar morphology shown here. (D) ICM isolated by immunosurgery was cultured in vitro for 4 days. Stat3-Independent Pluripotency Maintenance Factor 639 Figure 7. DNA Recognition and Proposed Functions of Nanog (A) Consensus binding sequence was determined by SELEX and shown with Sequence logo (Schneider and Stephens, 1990), which was generated by WebLogo (www.bio.cam.ac.uk/cgi-bin/seqlogo/ logo.cgi). The height of each letter is proportional to its frequency, and the letters are sorted so that the most common one is on top. The height of the entire stack is adjusted to signify the information content (measured in bits) of the sequence at that position. (B) 32P-labeled oligonucleotides containing the consensus sequence (wt: tgaggcccaccctCAGCCATTAaCctttttactttattct) or mutated sequence (mut: tgaggcccaccctCAGCCAGGAaCctttttactttattct) were incubated with nuclear extracts of F9 embryonic carcinoma cells, separated by SDS-PAGE, and visualized by a phosphoimager (Tokuzawa et al., 2003). Specificity of the reaction was tested by addition of excess amount of cold oligonucleotides. (C) Proposed functions of Nanog, Oct3/4, and Stat3 in preimplantation embryos (upper) and ES cells (lower). (Chambers et al., 2003 [Cell, this issue]). These data argue that Nanog is the missing fundamental factor that underlies pluripotency in both ICM and ES cells. The ability to support LIF/Stat3-independent selfrenewal is unique to Nanog. We identified eight other ecat genes that are expressed in ES cells as specifically as nanog and oct3/4. We have shown that ecat3 encoding the F box protein Fbx15 is a novel target of Oct3/4 and Sox2 (Tokuzawa et al., 2003). We have also demonstrated that ecat5 encoding a constitutively active Ras protein (ERas) plays an important role in tumor-like property of ES cells (Takahashi et al., 2003). However, we could not achieve LIF/Stat3-independet self-renewal with Ecat3, Ecat5, other Ecats, Oct3/4, or Sox2. These results highlighted the importance of Ecat4/Nanog in maintenance of pluripotency. Three lines of evidence indicate that Nanog functions independently of Stat3. First, phosphorylation kinetics of Stat3 is not elevated in CAG-nanog ES cells. It responded normally to LIF removal and stimulation. Second, loss-of- function phenotypes in ES cells are different between Stat3 and Nanog. Inactivation of the Stat3 pathway by LIF depletion or a dominant-negative mutant resulted in differentiation into multiple cell lineages including primitive endoderm and mesoderm (Niwa et al., 1998). However, terminal differentiation into parietal and/or visceral endoderm was not observed. Expression of gata4 and gata6 was barely increased by those treatments (Fujikura et al., 2002). In contrast, nanog null cells spontaneously differentiated into parietal and visceral endoderm expressing high levels of gata4 and gata6. Third, LIF was still able to enhance growth of CAG-nanog cells. If Nanog were a downstream target of Stat3, these results could not be explained. How does Nanog maintain self-renewal of ES cells? One of the mechanisms may be transcriptional repression of genes that promote differentiation, such as gata4 and gata6. We found that both gata4 and gata6 were upregulated in nanog null cells. Forced expression of gata4 or gata6 in ES cells was sufficient to induce extraembryonic endoderm differentiation (Fujikura et al., 2002). Expression patterns of marker genes were nearly identical between their gata6-overexpressing cells and our nanog null cells. Therefore, the upregulation of gata4 and gata6 could be sufficient to explain spontaneous differentiation into extraembryonic endoderm in nanog null cells. It has been shown that gata6 may be upstream of gata4, since inactivation of gata6 resulted in repression of gata4 (Morrisey et al., 1998) while inactivation of gata4 increased gata6 expression (Kuo et al., 1997). We found that the gata6 enhancer region (Molkentin et al., 2000; Sun-Wada et al., 2000) contains the DNA recognition motif of Nanog determined by SELEX, suggesting that Nanog might directly repress gata6, thereby inhibiting ES cell differentiation. In addition, the Nanog consensus sequence is found in the enhancer region of rex1 (BenShushan et al., 1998). Thus, Nanog may also regulate ES cell-specific genes to maintain pluripotency. Further studies, such as reporter gene assays and gel-mobility shift assays, are required to determine whether Nanog indeed binds to the consensus sequence in the gata6 or rex1 to regulate their expression. Another important issue is the relationship between Nanog and Oct3/4. The loss-of-function phenotypes of the two transcription factors are different. Disruption of nanog resulted in cell populations that expressed markers of both parietal and visceral endoderm and could be serially passaged for several months. In contrast, oct3/4 null cells differentiated into trophectoderm (Nichols et al., 1998; Niwa et al., 2000). Overexpression phenotypes are also different. Overexpression of nanog led to LIF-independent self-renewal of ES cells, while oct3/4 overexpression resulted in differentiation into multiple cell lineages, a similar phenotype caused by Stat3 inactivation (Niwa et al., 2000). These results indicate that the primary function of Oct3/4 is to prevent trophectoderm differentiation of ICM and ES cells. In contrast, Nanog independently prevents differentiation into extraembryonic endoderm and actively maintains pluripotency. Based on the finding of our present work and previous reports by others, we propose a model for the roles of Nanog, Oct3/4, and Stat3 in the maintenance of pluripo- Cell 640 tency (Figure 7C). During preimplantation development, the first cell fate determination takes place at morula stage, when cells remain pluripotent as ICM or differentiate into trophectoderm. At this stage, nanog expression is still low and Oct3/4 is the key determinant of cell fate. Cells that lost oct3/4 expression differentiate into trophectoderm. The second cell fate determination subsequently takes place at blastocyst stage when cells of ICM remain pluripotent as epiblast or differentiate into primitive endoderm. Nanog is expressed at this stage and functions as the crucial determinant of cell fate. Cells that express nanog remain pluripotent, while cells without nanog differentiate into primitive endoderm. ES cells derived from ICM require both Oct3/4 and Nanog to prevent differentiation into trophectoderm and primitive endoderm, respectively. However, these two transcription factors of normal expression level are not sufficient for ES cell self-renewal. Additional factor(s), such as Stat3 activated by LIF, are required to support prolonged maintenance of pluripotency. Increased expression of Oct3/4 does not render ES cells independent of LIF, but instead induces primitive endoderm differentiation. This suggests that the function of Oct3/4 in ES cells is to specify cell fate into either pluripotent cells or primitive endoderm by blocking trophectoderm differentiation. In contrast, our data demonstrated that elevated expression of Nanog bypasses the LIF/Stat3 pathway and maintains self-renewal of ES cells. Thus, Nanog not only blocks primitive endoderm differentiation, but also actively maintains pluripotency. In conclusion, our study revealed two important properties of the homeoprotein Nanog—the fundamental role in pluripotency of both ICM and ES cells, and the ability to maintain ES cell self-renewal without LIF. Oct3/4 and Stat3, the two well-known players in pluripotency, have only one of the two properties. Identification of critical regulatory genes such as nanog is a crucial step in understanding early embryogenesis and in exploiting pluripotent cells for therapeutic goals. Experimental Procedures Constitutive Expression of the ecat Genes in ES Cells The coding region of the ecat genes, oct3/4, or sox2 was introduced with Gateway technology (Invitrogen) into pPyCAGIP vector that allowed episomal expression in MG1.19 ES cells (Aubert et al., 2002; Niwa et al., 2002). Transfection into MG1.19 cells was performed with LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocol. Puromycin selection (2 g/ml) was continued throughout experiments. Targeted Disruption of the Mouse nanog Gene Two targeting vectors with promoter trap selection was designed to replace exon 2 of the mouse nanog gene, which contains the homeodomain, with an IRES (internal ribosome entry site)-geo cassette (Mountford et al., 1994) or the IRES-hygr cassette. A 4 kb fragment containing intron 1 and a 1.5 kb fragment containing exon 3, intron 3, and exon 4 were amplified from mouse genomic DNA and used as the 5⬘ and 3⬘ homologous regions of the targeting vector. The 5⬘ arm was amplified by expand long template PCR system (Roche) with primers S4 (AGGGTCTGCTACTGAGATGCTC TG) and AS4 (AGGCAGGTCTTCAGAGGAAGGGCG). The 3⬘ arm was amplified with primes IntS3.2 (CGGGCTGTAGACCTGTCTGCATTC TG) and RACE2 (GGTCCTTCTGTCTCATCCTCGAGAGT). Either the geo or hygr cassette was ligated between the two DNA fragments. The resulting targeting vectors were linearized with SacII digestion and introduced into RF8 ES cells by electroporation (Meiner et al., 1996). Genomic DNAs from G418- or hygromycin B-resistant colonies were screened for homologous recombination by PCR and Southern blot. Analyses of Marker Gene Expression Total RNA was isolated with Trizol (Invitrogen). Northern blot analyses were performed with formalin-agarose gel as previously described (Yamanaka et al., 2000, 1998). For RT-PCR, first strand cDNA was synthesized with ReverTra Ace (Toyobo, Japan), and PCR was carried out with ExTaq (Takara, Japan). Most PCR primers were described by others (Fujikura et al., 2002). Other primers used are 6047-S4 (AGGGTCTGCTACTGAGATGCTCTG) and 6047-AS5 (CAAC CACTGGTTTTTCTGCCACCG) for nanog, Oct3/4-U474 (CTGAGGG CCAGGCAGGAGCACGAG) and Oct3/4- L935 (CTGTAGGGAGGGC TTCGGGCACTT) for oct3/4, NAT1-U283 (ATTCTTCGTTGTCAAGC CGCCAAAGTGGAG) and NAT1-L476 (AGTTGTTTGCTGCGGAGTTG TCATCTCGTC) for nat1, Cdx2-S775 (GGCGAAACCTGTGCGAGTG GATGCGGAA) and Cdx2-AS1210 (GATTGCTGTGCCGCCGCCGCT TCAGACC) for cdx2, LimB.S354 (CTGTTCTGAAAGTGAATGTGGTG GCCCC) and LamB-AS945 (GTTTAATCGCCTTCTCTGCTGCAAC CTG) for lamininB1, and TM-S1071 (CCAGGCTCTTACTCCTGTA) and TM-AS1300 (TGGCACTGAAACTCGCAGTT) for thrombomodulin. Isolation of ICM by Immunosurgery Blastocysts were incubated for 1 min at room temperature in acidic Tyrode’s solution (Sigma) to remove Zona Pellucida and washed three times with M16 medium (Sigma). Zona-free embryos were incubated for 10 min at 37⬚C with ␣-mouse antiserum (diluted 20 times in M16 medium), rinsed three times with M16, and incubated for 30 minutes at 37⬚C with guinea pig complement (ICN Biomedicals, Inc., diluted 20 times in M16 medium). After washing three times with M16 medium, ICM were isolated from dead trophectoderm and cultured in gelatin-coated plate with ES cell medium. SELEX The coding region of the mouse nanog gene was introduced into pIH1119 to produce a fusion protein consisting of MBP and Nanog. The fusion was induced in E. coli BL21AI (Invitrogen) and was purified with amylose-beads (New England Biolabs). SELEX was performed as follows. Double strand oligonucleotide was synthesized in reaction mixture containing 1 g SELEX-N20-Oligo (TAGGCATGT GGATCCGTCTGGCN20GCGTACCGGATCCACCTTCGAT), 1 g SELEX-N20RV primer (ATCGAAGGTGGATCCGGTACGC), 2.5 l 10⫻ Klenow buffer, 4 l 10 mM dNTP, and 14.7 l distilled water. The mixture was denatured at 95⬚C for 5 min and annealed at 54⬚C for 5 min. Klenow fragment (0.225 U in 3.6 l of 1⫻ Klenow buffer) was added to the mixture, which was then incubated at 25⬚C for 40 min. The double-strand DNA was purified by phenol/chloroform extraction and ethanol precipitation and resuspended in 20 l distilled water. DNA-protein binding was performed at 4⬚C for 30 min in reaction mixture consisting of 20 l DNA, 30 l MBP-Nanogbound beads, 20 l of 5⫻ binding buffer (100 mM HEPES-KOH [pH 7.9], 1 mM EDTA, 1 M KCl, and 50% Glycerol), and 30 l of water. The beads were washed six times with 100 l lysis buffer (20 mM Tris-HCl [ph 7.4], 200 mM NaCl, and 1mM EDTA) supplemented with 1 mM DTT and 0.2 mM PMSF. DNA was purified by phenol/ chloroform extraction and ethanol precipitation and resuspended in 20 l distilled water. 5 l of this was used for PCR amplification in reaction buffer containing 5 l of 10⫻ ExTaq buffer, 4 l of 2.5 mM dNTP, 1 l of 30 M SELEX-N20FW primer (TAGGCATGTGG ATCCGTCTGGC), 1 l of 30 M SELEX-N20RV primer, 0.25 L of ExTaq polymerase, and 33.75 L of water. PCR program consisted of the initial denaturation at 95⬚C for 5 min, 20 cycles of 95⬚C for 1 min, 54⬚C for 1.5 min, and 72⬚C for 1 min, and the final extension at 72⬚C for 7 min. Amplified products were purified by Micro-bio spin column (BioRad) and concentrated to 20 l by ethanol precipitation, of which half was applied to another selection round. This procedure was repeated five times for enrichment. Amplified products in the final reaction were ligated into pCR2.1 vector (Invitrogen). Thirty clones were randomly selected and sequenced. Stat3-Independent Pluripotency Maintenance Factor 641 Acknowledgments to regulate endodermal-specific promoter expression. Proc. Natl. Acad. Sci. USA 99, 3663–3667. We thank Drs. Ian Chambers, Jennifer Nichols, and Austin Smith for sharing data prior to publication. We thank Tomoko Ichisaka and Yukiko Samitsu for blastocyst microinjection, Junko Iida and Chihiro Takigawa for technical assistance, and Takashi Iwasaki for histological analyses. We also thank Dr. Hung Li for agreeing with the nomenclature. We are grateful to Dr. Hitoshi Niwa for providing various materials essential for this study. This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; Uehara Memorial Foundation; and Sumitomo Research Foundation (to S.Y.). Hanna, L.A., Foreman, R.K., Tarasenko, I.A., Kessler, D.S., and Labosky, P.A. (2002). Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 16, 2650–2661. Received: December 9, 2002 Revised: April 29, 2003 Accepted: April 30, 2003 Published: May 29, 2003 References Aubert, J., Dunstan, H., Chambers, I., and Smith, A. (2002). Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 20, 1240–1245. Avilion, A.A., Nicolis, S.K., Pevny, L.H., Perez, L., Vivian, N., and Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140. Ben-Shushan, E., Thompson, J.R., Gudas, L.J., and Bergman, Y. (1998). Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18, 1866–1878. Berger, C.N., and Sturm, K.S. (1997). Self renewal of embryonic stem cells in the absence of feeder cells and exogenous leukaemia inhibitory factor. Growth Factors 14, 145–159. Burdon, T., Smith, A., and Savatier, P. (2002). Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12, 432–438. Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., and Smith, A. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, this issue. Colman, A., and Burley, J.C. (2001). A legal and ethical tightrope. Science, ethics and legislation of stem cell research. EMBO Rep. 2, 2–5. Dani, C., Chambers, I., Johnstone, S., Robertson, M., Ebrahimi, B., Saito, M., Taga, T., Li, M., Burdon, T., Nichols, J., and Smith, A. (1998). Paracrine induction of stem cell renewal by LIF-deficient cells: a new ES cell regulatory pathway. Dev. Biol. 203, 149–162. Ernst, M., Oates, A., and Dunn, A.R. (1996). Gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogen-activated protein kinase pathways. J. Biol. Chem. 271, 30136–30143. Ernst, M., Novak, U., Nicholson, S.E., Layton, J.E., and Dunn, A.R. (1999). The carboxyl-terminal domains of gp130-related cytokine receptors are necessary for suppressing embryonic stem cell differentiation. Involvement of STAT3. J. Biol. Chem. 274, 9729–9737. Evans, M.J., and Kaufman, M.H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. Fan, Y., Melhem, M.F., and Chaillet, J.R. (1999). Forced expression of the homeobox-containing gene Pem blocks differentiation of embryonic stem cells. Dev. Biol. 210, 481–496. Fujikura, J., Yamato, E., Yonemura, S., Hosoda, K., Masui, S., Nakao, K., Miyazaki Ji, J., and Niwa, H. (2002). Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16, 784–789. Gassmann, M., Donoho, G., and Berg, P. (1995). Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 92, 1292–1296. Guo, Y., Costa, R., Ramsey, H., Starnes, T., Vance, G., Robertson, K., Kelley, M., Reinbold, R., Scholer, H., and Hromas, R. (2002). The embryonic stem cell transcription factors Oct-4 and FoxD3 interact Harvey, R.P. (1996). NK-2 homeobox genes and heart development. Dev. Biol. 178, 203–216. Ihle, J.N. (1996). STATs: signal transducers and activators of transcription. Cell 84, 331–334. Jackson, M., Baird, J.W., Cambray, N., Ansell, J.D., Forrester, L.M., and Graham, G.J. (2002). Cloning and characterization of Ehox, a novel homeobox gene essential for embryonic stem cell differentiation. J. Biol. Chem. 277, 38683–38692. Kuo, C.T., Morrisey, E.E., Anandappa, R., Sigrist, K., Lu, M.M., Parmacek, M.S., Soudais, C., and Leiden, J.M. (1997). GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11, 1048–1060. Lachmann, P. (2001). Stem cell research—why is it regarded as a threat? An investigation of the economic and ethical arguments made against research with human embryonic stem cells. EMBO Rep. 2, 165–168. Li, E., Bestor, T.H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. Li, M., Sendtner, M., and Smith, A. (1995). Essential function of LIF receptor in motor neurons. Nature 378, 724–727. Martin, G.R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638. Matsuda, T., Nakamura, T., Nakao, K., Arai, T., Katsuki, M., Heike, T., and Yokota, T. (1999). STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18, 4261–4269. Meiner, V.L., Cases, S., Myers, H.M., Sande, E.R., Bellosta, S., Schambelan, M., Pitas, R.E., McGuire, J., Herz, J., and Farese, R.V., Jr. (1996). Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc. Natl. Acad. Sci. USA 93, 14041–14046. Mieth, D. (2000). Going to the roots of the stem cell debate. The ethical problems of using embryos for research. EMBO Rep. 1, 4–6. Molkentin, J.D., Antos, C., Mercer, B., Taigen, T., Miano, J.M., and Olson, E.N. (2000). Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev. Biol. 217, 301–309. Morrisey, E.E., Tang, Z., Sigrist, K., Lu, M.M., Jiang, F., Ip, H.S., and Parmacek, M.S. (1998). GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12, 3579–3590. Mountford, P., Zevnik, B., Duwel, A., Nichols, J., Li, M., Dani, C., Robertson, M., Chambers, I., and Smith, A. (1994). Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91, 4303–4307. Nakajima, K., Yamanaka, Y., Nakae, K., Kojima, H., Ichiba, M., Kiuchi, N., Kitaoka, T., Fukada, T., Hibi, M., and Hirano, T. (1996). A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15, 3651–3658. Nichols, J., Chambers, I., Taga, T., and Smith, A. (2001). Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128, 2333–2339. Nichols, J., Evans, E.P., and Smith, A.G. (1990). Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 110, 1341–1348. Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., Scholer, H., and Smith, A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391. Niwa, H., Burdon, T., Chambers, I., and Smith, A. (1998). Self-renewal Cell 642 of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060. target of Oct3/4 but is dispensable for embryonic stem cell selfrenewal and mouse development. Mol. Cell. Biol. 23, 2699–2708. Niwa, H., Miyazaki, J., and Smith, A.G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or selfrenewal of ES cells. Nat. Genet. 24, 372–376. Wang, S.H., Tsai, M.S., Chiang, M.F., and Li, H. (2003). A novel NKtype homeobox gene, ENK (early embryo specific NK), preferentially expressed in embryonic stem cells. Gene Expr. Patterns 3, 99–103. Niwa, H., Masui, S., Chambers, I., Smith, A.G., and Miyazaki, J. (2002). Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct3/4 in embryonic stem cells. Mol. Cell. Biol. 22, 1526–1536. Ware, C.B., Horowitz, M.C., Renshaw, B.R., Hunt, J.S., Liggitt, D., Koblar, S.A., Gliniak, B.C., McKenna, H.J., Papayannopoulou, T., Thoma, B., et al. (1995). Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 121, 1283–1299. Okamoto, K., Okazawa, H., Okuda, A., Sakai, M., Muramatsu, M., and Hamada, H. (1990). A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472. Okuda, A., Fukushima, A., Nishimoto, M., Orimo, A., Yamagishi, T., Nabeshima, Y., Kuro-o, M., Nabeshima, Y., Boon, K., Keaveney, M., et al. (1998). UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 17, 2019-2032. Palmieri, S.L., Peter, W., Hess, H., and Scholer, H.R. (1994). Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166, 259–267. Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R.C., and Melton, D.A. (2002). “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600. Raz, R., Lee, C.K., Cannizzaro, L.A., d’Eustachio, P., and Levy, D.E. (1999). Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 96, 2846–2851. Reubinoff, B.E., Pera, M.F., Fong, C.Y., Trounson, A., and Bongso, A. (2000). Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404. Rogers, M.B., Hosler, B.A., and Gudas, L.J. (1991). Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113, 815–824. Schneider, T.D., and Stephens, R.M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097– 6100. Scholer, H.R., Dressler, G.R., Balling, R., Rohdewohld, H., and Gruss, P. (1990). Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 9, 2185–2195. Smith, A. (1998). Cell therapy: in search of pluripotency. Curr. Biol. 8, R802–R804. Smith, A.G., Heath, J.K., Donaldson, D.D., Wong, G.G., Moreau, J., Stahl, M., and Rogers, D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690. Stewart, C.L., Kaspar, P., Brunet, L.J., Bhatt, H., Gadi, I., Kontgen, F., and Abbondanzo, S.J. (1992). Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359, 76–79. Sun-Wada, G.H., Manabe, S., Yoshimizu, T., Yamaguchi, C., Oka, T., Wada, Y., and Futai, M. (2000). Upstream regions directing heartspecific expression of the GATA6 gene during mouse early development. J. Biochem. (Tokyo) 127, 703–709. Takahashi, K., Mitsui, K., and Yamanaka, S. (2003). Role of ERas in tumor-like properties in mouse embryonic stem cells. Nature 423, 541–545. Takeda, K., Noguchi, K., Shi, W., Tanaka, T., Matsumoto, M., Yoshida, N., Kishimoto, T., and Akira, S. (1997). Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94, 3801–3804. Thomson, J.A., Itskovitz-Eldor, J., Shapiro, S.S., Waknitz, M.A., Swiergiel, J.J., Marshall, V.S., and Jones, J.M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145– 1147. Tokuzawa, Y., Kaiho, E., Maruyama, M., Takahashi, K., Mitsui, K., Maeda, M., Niwa, H., and Yamanaka, S. (2003). Fbx15 is a novel Williams, R.L., Hilton, D.J., Pease, S., Willson, T.A., Stewart, C.L., Gearing, D.P., Wagner, E.F., Metcalf, D., Nicola, N.A., and Gough, N.M. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687. Yamanaka, S., Zhang, X.Y., Miura, K., Kim, S., and Iwao, H. (1998). The human gene encoding the lectin-type oxidized LDL receptor (OLR1) is a novel member of the natural killer gene complex with a unique expression profile. Genomics 54, 191–199. Yamanaka, S., Zhang, X.Y., Maeda, M., Miura, K., Wang, S., Farese, R.V., Jr., Iwao, H., and Innerarity, T.L. (2000). Essential role of NAT1/ p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 19, 5533–5541. Yoshida, K., Taga, T., Saito, M., Suematsu, S., Kumanogoh, A., Tanaka, T., Fujiwara, H., Hirata, M., Yamagami, T., Nakahata, T., et al. (1996). Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc. Natl. Acad. Sci. USA 93, 407–411. Yuan, H., Corbi, N., Basilico, C., and Dailey, L. (1995). Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9, 2635–2645. Accession Numbers The GenBank accession number for the mouse nanog cDNA sequence reported in this paper is AB093574.