Computer lab Enzyme kinetics and characterization of reaction intermediates
advertisement

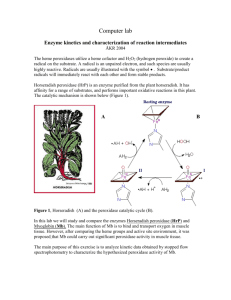
Computer lab Enzyme kinetics and characterization of reaction intermediates ÅKR 2003 The heme peroxidases utilize a heme cofactor and H2O2 (hydrogen peroxide) to create a radical on the substrate. A radical is an unpaired electron, and such species are usually highly reactive. Radicals are usually illustrated with the symbol . Substrate/product radicals will immediately react with each other and form stable products. Horseradish peroxidase (HrP) is an enzyme purified from the plant horseradish. It has affinity for a range of substrates, and performs important oxidative reactions in this plant. The catalytic mechanism is shown below (Figure 1). A Figure 1, Horseradish (A) and the peroxidase catalytic cycle (B). In this lab we will study and compare the enzymes Horseradish peroxidase (HrP) and Myoglobin (Mb). The main function of Mb is to bind and transport oxygen in muscle tissue. However, after comparing the heme groups and active site environment, it was proposed that Mb could carry out significant peroxidase activity in muscle tissue. The main purpose of this exercise is to analyze kinetic data obtained by stopped flow spectrophotometry to characterize the hypothesized peroxidase activity of Mb. B Summary of experiments: 1) Determine the Vmax and kcat for HrP and Mb at pH = 6.5 and T = 20 C using different concentrations of the substrate ortho-phenylenediamine (OPD). 2) Investigate the rapid stopped flow kinetics (in ms time scale) for the reaction of enzyme and H2O2. 3) Discuss which properties that influence the catalytic efficiency. NB! SAVE ALL FIGURES YOU MAKE DURING THE EXERCISES Determination of kinetic constants for HrP and Mb using OPD as substrate Ortho-phenylenediamine is an excellent substrate for HrP, and because a colorless substrate is converted to an orange-brown product, the reaction progress can easily be monitored by spectrophotometry. We will use the Michaelis –Menten formalism to extract the Vmax parameter and subsequently calculate the turnover number, kcat, that describes the efficiency of the enzymes. Description of Michaelis – Menten experiment: 1) 2) 3) 4) Import data from CSV files. Determine initial rates from the absorption at 450 nm. Make a Michaelis-Menten plot (initial rate versus [OPD]). Fit data, calculate kcat and answer questions. Import of data Start Origin 7.0. File -> Import -> ASCII Options Do this only once? You may have to do it each time due to user restrictions Press Update Options, then File ->Save Template As -> OK If you want to import a single data set press If you want to import multiple data sets press NB! To import a single data set, select one worksheet. Before importing multiple data sets, close all worksheets! Format of imported data Click to select column with wavelength data (nm) Click to select column with absorbance data (AU) Click to select column with wavelength data (nm) Click to select row with time data (s) Click to select row with absorbance data at a single wavelength (AU) Remember, by right clicking a cell, column, or row, you can either insert or delete an item. Example: To isolate absorbance values only, delete the first row and first column. Exercise 1 Make a 3D plot from data showing the production of DAP and calculate the extinction coefficient of this product: 1) 2) 3) 4) Select an imported data set (HRP_0100.CSV) Delete row 1 and column 1 from the imported data. To convert row to column press: Edit -> Transpose -> Yes While imported data window is selected, press: Edit -> Convert to matrix -> Direct. A yellow matrix containing all absorption data turns up. 5) Then set the matrix dimensions, press: Matrix -> Dimensions 8) Set X axis as wavelength (low- high) 9) Set Y axis as time in seconds (start --- end) 10) Plotting: While matrix window is selected press: Plot, and then 3D Color fill surface, to look at mesh surface 3D X constant with base, to look at time lines 3D Y constant with base, to look at single absorption spectra. Use this toolbar to manipulate view Use this button to read values from plot (navigate with keyboard arrows) At which wavelength is the product (DAP) absorption maximum found? To calculate the extinction coefficient open the Project “epsilondetermination.opj ”. Plot the data in the worksheet. This is an spectrum recorded when all substrate have been converted to product. The initial substrate concentration was 0.1 mM. Calculate the extinction coefficient 450 (mM-1cm-1) of the product DAP at 450 nm by using Beers law; A450 = 450 [DAP] 1 cm . Remember that 2 OPD DAP. What is the extinction coefficient 450 of DAP? Why do we use the last spectrum to calculate extinction coefficient? Exercise 2 Determination of Vmax and kcat Different concentrations of substrate have been added to Mb and HrP. Stopped flow data have been recorded and stored as CVS files. Data files HrP [OPD] mM 0.05 0.1 0.15 0.2 0.3 0.5 0.8 1.2 Mb Filename HRP_0050 HRP_0100 HRP_0150 HRP_0200 HRP_0300 HRP_0500 HRP_0800 HRP_1200 [OPD] mM 0.02 0.03 0.05 0.15 0.2 0.3 - Filename MB_0020 MB_0030 MB_0050 MB_0150 MB_0200 MB_0300 - Find initial rates (v0) for all reactions Import data file (see procedure above). Create a new worksheet. Copy first row of imported data into new worksheet (time). Then find the row containing data at 450 nm. Copy this row into the second row of the new worksheet. To create X and Y columns for plotting press: Edit -> Transpose, and click Yes. Delete the first row (select row, right click and press delete). Select X and Y column and press the button. Is the curve linear? If not, select only 6-10 points at the beginning of the curve (skip the first 1-3 points) and plot them again. To find the initial rate do a linear fit of the first data points: Analyzis -> Fit linear (the result of the fit shows up in the box at the lower right corner of the screen, scroll and find the slope B). V0 = B/ 0.5 * 450, DAP Calculate v0 for all data sets, and write them down. Michaelis- Menten plot For each protein, create a worksheet with [OPD] in the X-column and v0 in the Y-column. Plot each worksheet with and fit using the Advanced fitting tool. Press: Analysis -> Non linear curve fit -> Advanced fitting tool. A new dialog box appers: Press: Function -> Select Choose: Category = Pharmacology Function = OneSiteBind This function resembles the Michaelis –Menten equation: B = Vmax K = Km X = [OPD] Press: Action -> Fit, Active dataset. Choose all initial fitting parameters = 1 and press 100 Iter. Write down the best fitting parameters. The enzyme concentrations used are [HrP] = 1.0 *10-5 mM and [Mb] = 5.8 *10-3 mM For both HrP and Mb, calculate kcat = Vmax / [Enzyme] What does the kcat constants tell you about the of the reactions? How many times faster is HrP than Mb catalyzing the reaction 2 OPD DAP ? Exercise 3 Introduction Rapid stopped flow kinetics detection of intermediates The reaction cycle of HrP is shown in Figure 1. Two intermediates, compound I (A) and compound II (B), can be detected when the enzyme reacts with H2O2. A B In this experiment the ferric (Fe3+) form of the two enzymes HrP and Mb are allowed to react with hydrogen peroxide (H2O2) in absence of the substrate OPD. We want to study the reaction of H2O2 with the two enzymes HrP and Mb. Perhaps we can gain some insight that will explain the difference in catalytic efficiency between the two enzymes? Soret () band and bands 350 400 450 500 550 600 650 700 Wavelenght (nm) Figure 2. Visible spectrum of ferric (Fe3+) HrP p p* (, Low intensity) p p* (, High intensity) p p* (, Low intensity) 350 400 450 500 550 600 650 Charge Transfer Low intensity 700 Wavelength Wavelength (nm) 450 500 550 600 650 700 Wavelength Figure 3. Visible spectrum of ferric (Fe3+) Mb Experimental The data files used in this exercise are stored in the catalogs “Rapid_kinetics_HrP” and “Rapid_kinetics_HrP”. 1) Make a plot where you compare the “resting” ferric HrP with the following stopped flow data (2D-graph). The interesting region is 450 – 700 nm. 0 s (Hrpresting.opj) 75 ms (HrPH20215s.CSV, first spectrum) 15 s (HrPH20215s.CSV, last spectrum) 2) Make similar plot for Mb 0 s (Hrpresting.opj) 5 ms (MbH2O2.CSV, first spectrum) 500 ms (MbH2O2.CSV, last spectrum) A) From the plot for HrP at different times, make a table of the absorbance values at 496 nm, 543 nm, 578 nm and 650 nm at 0 s, 75 ms, and 15 s. Are there any peaks that first seem to increase in the first time interval (0 s – 75 ms) and then decrease in the second time interval (75 ms – 15 s)? Which spectrum corresponds to which intermediate in Figure 1? * Make a 3D plot for HrPH202.CSV and for HrPH20215s.CSV. For each wavelength, calculate first order rate constants k using: Analysis -> Fit exponential decay -> First order. TIPS!! If the fitting is bad (e.g. t1 > 10 ), use the Advanced fitting tool, select the exponential category and use the function ExpDec1 with the parameter A1 < 0. Another trick if the fitting is bad is to exclude data that does not “look exponential” or restart the Origin program. The rate constant k = 1/ t1 (compare the function used by Origin to the integrated first order function at page 7, line 4 in the compendium from A.V. Agasøster). A high rate constant indicates a fast reaction. Insert the calculated rate constants in the table below. Wavlelength 5 – 2000 ms 75 ms – 15 s First Order Rate Constants for HrP 496 nm 543 nm 578 nm 650 nm B) Repeat the procedure above for Mb. Since the Mb + H2O2 reaction is finished in 500 ms, there is only one data file (MbH2O2.CSV). Skip the question above marked with “*” and answer this one instead: In the Mb + H2O2 reaction only one of the intermediates in Figure 1 is observed. Which one? (Hint: look for similar spectral features). Make a 3D plot for MbH2O2.CSV as well. Wavlelength 5 – 500 ms First Order Rate Constants for Mb 503 nm 550 nm 588 nm 640 nm Both compound I and compound II are very reactive. Consider the results from the Michaelis- Menten experiments (calculation of the turnover number kcat) and the rapid stopped flow studies of reaction intermediates in the absence of substrate. Discuss the seemingly contradictory results. What do you think determine the formation of the product DAP from OPD? Considering the low substrate affinity of H2O2 activated Mb; do you think Mb can carry out significant peroxidase activity in human tissue?