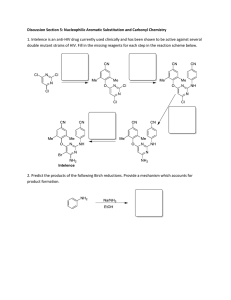
Doctor of Philosophy for the degree of
advertisement

AN ABSTRACT OF THE THESIS OF Hye-Dong Yoo for the degree of Doctor of Philosophy in Pharmacy presented on October 13. 1997 Title: Bioactive Secondary Metabolites from Marine Algae and Study of Oxidized Anandamide Derivatives Redacted for Privacy Abstract approved: William H. Gerwick My investigations of the natural products of marine algae have resulted in the discovery of several new secondary metabolites. Bioassay-guided fractionation led to the isolation of these new compounds and spectroscopic analysis was utilized in their structural characterization. Two new and potent antimitotic metabolites, curacins B and C, were isolated from a Curacao collection of Lyngbya majuscula. In addition, four curacin A analogs were prepared by semisynthetic methods. The structures of the new curacins and the curacin A analogs were determined by spectroscopic analysis in comparison with curacin A. The biological properties of the new natural products and synthetic derivatives of curacin A were examined. Investigations of another Curacao collection of L. majuscula revealed a new cytotoxic lipopeptide, microcolin C. Microcolin C was found to have an interesting profile of cytotoxicity to human cancer-derived cell lines. A new metabolite, vidalenolone, was isolated from an Indonesian red alga Vidalia sp. The structure of this new cyclopentenolone-containing compound was determined by a combination of spectroscopic methods. Filamentous cells isolated from female gametophytes of the brown alga Laminaria saccharina were cultured in flasks or bioreactors. These cultures produced a variety of w6-lipoxygenase metabolites: 13-hydroxy-9,11­ octadecadienoic acid (13-HODE), 13-hydroxy-6,9,11,15-octadecatetraenoic acid (13-HODTA), and 15-hydroxy-5,8,11,13-eicosatetraenoic acid (15-HETE). Five oxidized anandamide derivatives were prepared from anandamide through autoxidation in an exploration of ligand binding to the cannabinoid receptor. Their structures were determined by a combination of NMR spectroscopy and GC-MS. The cannabinoid receptor binding affinity of these derivatives was evaluated. This study revealed the following trend in activity: anandamide > 15- > 9- > 8- > 11- > 5-hydroxyanandamide. Copyright by Hye-Dong Yoo October 13, 1997 All Rights Reserved Bioactive Secondary Metabolites from Marine Algae and Study of Oxidized Anandamide Derivatives by Hye-Dong Yoo A THESIS submitted to Oregon State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy Completed October 13, 1997 Commencement June 1998 Doctor of Philosophy thesis of Hye-Dong Yoo presented on October 13, 1997 APPROVED: Redacted for Privacy Major Professor, representing Pharmacy Redacted for Privacy Dean of the College of Pharmacy Redacted for Privacy Dean of Grad e School I understand that my thesis will become part of the permanent collection of Oregon State University libraries. My signature below authorizes release of my thesis to any reader upon request. Redacted for Privacy Hye-Dong Yoo, Author ACKNOWLEDGEMENTS I wish to thank my major advisor, Dr. William H. Gerwick, for his generous support and guidance throughout my graduate studies. His unlimited patience and mentorship led me to completion of this project. I must thank my labmate Mary Ann Roberts for teaching me about algae and for encouragement during my research. I would like to thank Dr. George Constantine for his thoughtful advice and encouragement; Dr. Max Deinzer and Dr. Michael Hyman for accepting to be on my committee and their time; Dr. Mark Zabriskie and Dr. Philip Proteau for their insightful suggestions; Dr. Mitchell L. Wise for a long time friendship; Dr. Dale Nagle for his assistance in my early research years; Dr. Yongfu Li and Dr. Anna Boulanger for helpful discussions; Brian Arbogast for his mass spectral work; Roger Kohnert for NMR spectroscopy. I am also indebted to Dr. Greg Hooper, Mary Roberts, Brian Marquez, Lik Tan Tong and Namthip Sitachitta for their assistance with this manuscript. I would like to acknowledge Dr. Pete K. Freeman, Dr. Henry Schaup, and Dr. Christian Haugen for their unconditional support and friendship. My past and present labmates are also appreciated for their support and helpful disscussions. Finally, I wish to express my gratitude to my family for their patience and support. Funding for the work in this thesis was supported by the National Cancer Institute and Oregon Sea Grant. CONTRIBUTION OF AUTHORS Chapters V and VI of my thesis were collaborative efforts performed with two research groups. Dr. Greg Rorrer research group in the Department of Chemical Engineering was involved in bioreactor design and start-up and cultivation of brown alga, Laminaria saccharina female gametophyte cells in chapter V. Ken Soderstrom in the laboratory of Dr. Thomas Murray was responsible for the competitive receptor ligand displacement assay with a variety of hydroxyanandamides for Chapter VI. TABLE OF CONTENTS Page CHAPTER I. GENERAL INTRODUCTION 1 MARINE MICROORGANISMS BIOACTIVE METABOLITES FROM CYANOBACTERIA OXYLIPINS FROM MARINE ALGAE CHAPTER II. CURACINS, ANTIMITOTIC NATURAL PRODUCTS FROM THE MARINE CYANOBACTERIUM LYNGBYA MAJUSCULA 23 ABSTRACT 23 INTRODUCTION 24 RESULTS AND DISCUSSION 27 EXPERIMENTAL 50 CHAPTER III. MICROCOLINS, NEW ANTINEOPLASTIC NATURAL PRODUCTS FROM THE MARINE CYANOBACTERIUM LYNGBYA MAJUSCULA 59 ABSTRACT 59 INTRODUCTION 60 RESULTS AND DISCUSSION 63 EXPERIMENTAL 76 CHAPTER IV. VIDALENOLONE, A NOVEL SECONDARY METABOLITE FROM THE INDONESIAN RED MARINE ALGA VIDALIA SR 79 ABSTRACT 79 INTRODUCTION 80 RESULTS AND DISCUSSION 82 EXPERIMENTAL 89 CHAPTER V. ANALYSIS OF OXYLIPIN PRODUCTS FROM GAMETOPHYTE CELL CULTURE OF BROWN ALGA LAMINARIA SACCHARINA 91 ABSTRACT 91 INTRODUCTION 92 RESULTS AND DISCUSSION 97 EXPERIMENTAL 112 CHAPTER VI. PRODUCTION OF OXIDIZED ANANDAMIDE DERIVATIVES AND EVALUATION OF THEIR BINDING AFFINITY TO THE CANNABINOID RECEPTOR (CB1) 115 ABSTRACT 115 INTRODUCTION 116 RESULTS AND DISCUSSION 118 EXPERIMENTAL 132 CHAPTER VII. CONCLUSION 135 BIBLIOGRAPHY 138 APPENDIX 146 List of Figures Figure Page 1.1 Biosynthesis of the products of arachidonic acid in mammals. 15 11.1 Structures of selected secondary metabolites from Lyngbya majuscula. 25 Mean graphs of curacins B and C (95:5) mixture obtained from the National Cancer Institute 60 cell line in vitro screen. 28 11.3 Flow chart of fractionation of Lyngbya majuscula. 29 11.4 Comparison of 1H NMR spectra of curacins A, B, and C. 30 11.5 Geometrical configuration of conjugated diene of curacin B (14) determined by NOESY in CDCI3. 33 11.6 Thermal isomerization of curacins A, B, and C. 34 11.7 GC-MS traces for thermal isomeration of curacins A, B, and C. 35 11.8 Natural antimitotic agents and synthetic derivatives of the curacins. 37 11.2 11.9 11-1 NMR spectrum of cyclopropyl ring opened curacin A (18) in C6D6. 39 1H-1H COSY spectrum of cyclopropyl ring opened Curacin A (18) in C6D6. 40 11.11 1H NMR spectrum of 19-epi curacin A (19) in C6D6. 41 11.12 1H-1H COSY of 19-epi curacin A (19) in C6D6. 42 11.13 1H NMR spectrum of thiazoline ring opened curacin A 11.10 (20) in C6D6. 43 11.14 HMBC plot of thiazoline ring opened curacin A (20) in C6D6. 11.15 1H NMR spectrum of derivative 21 in C6D6. 45 11.16 'H NMR spectrum of curadisulfide A (22) in C6D6. 46 44 List of Figures (continued) Figure Page Flow chart for the isolation of microcolins. 64 111.2 Structures of microcolins A-D (7-10). 66 111.3 Mass fragmentations for microcolin isomers. 67 111.4 Comparison of 11-1 NMR spectra of microcolins A, B, and C (7-9) at 300 MHz in CDCI3. 69 111.5 TOCSY contour plot of microcolin C (9) at 600 MHz (CDCI3). 70 111.6 Enzymatic hydrolysis of microcolin B. 72 111.7 1H spectrum of microcolin D (10) at 600 MHz (CDCI3). 74 111.8 1H-1H COSY contour plot of microcolin D (10) at 600 MHz 111.1 (CDCI3). 75 IV.1 1H NMR spectrum of vidalenolone (13). 83 IV.2 Partial structures of vidalenolone (13). 85 IV.3 Structure of vidalenolone (13). 85 IV.4 Proposed biogenetic pathway of vidalenolone (13). 88 V.1 Expected fragment patterns of various oxylipins. 93 V.2 Overview of the identification and analysis of oxylipins in L. saccharina cell culture. 98 V.3. Total Ion Chromatogram (TIC) of TMS-hydroxy fatty acids produced from L. saccharina gametophyte cell suspension. V.4 V.5 99 Mass spectra and structures of TMS ether derivatives of methyl 13-HODTA (2) and methyl 15-HETE (3). 103 Mass spectra and structures of TMS ether derivatives of (A) methyl 13 -NODE (1) and (B) of co-eluted methyl ricinelaidic acid standard (4) and methyl 13 -NODE. 104 List of Figures (continued) Figure V.6 Page Hydroxy fatty acid production from precursors by 15-LOX/ peroxidase metabolism. 106 V.7 Yields of 13-HODTA from cultures added with different precursors. 108 V.8 Yields of 13-HODE from cultures added with different precursors. 109 V.9 Yields of 15-HETE from cultures added with different precursors. 110 V.10 Comparison of the yields of three oxylipins from feeding experiment. VI.1 Preparation and purification of various hydroxyanandamides. VI.2 Structures of various hydroxyanandamides. VI.3 Mass spectra of (A)15-hydroxyanandamide, (B) 11-hydroxyanandamide, (C) 9-hydroxyanandamide, (D) 8-hydroxyanandamide. 121 VI.4 Proposed structures of the ions at m/z 315, 327, 355, 407 formed due to rearrangement process corresponding to 8-(A), 9-(B), 11-(C), and 15-hydroxyanandamides (D), respectively. 122 VI.5 11-1 NMR spectrum of 15-hydroxyanandamide (3). 123 VI.6 1H-1H COSY of 15-hydroxyanandamide (3). 124 VI.7 1H NMR data assignments for 15-(3) and 5­ hydroxyanandamide (7). 125 VI.8 1H NMR spectrum of 5-hydroxyanandamide (7). 126 V1.9 1H-1H COSY of 5-hydroxyanandamide (7). 127 VI.10 Inhibition of the specific binding of [3H]CP-55940 to rat neuronal membrane by anandamide and oxidized anandamide derivatives. 131 List of Tables Figure Page 11.1 1H and 13C NMR data for curacins A (13), B (14), and C (15). 31 11.2 Biological activities of curacins on cell growth, tubulin polimerization, coichicine binding, and brine shrimp toxicity. 49 'H and 13C NMR data for 9 and 10. 71 Tumor cell growth inhibitory activity and cytotoxicity of microcolin C in the National Cancer Institute in vitro 60-cell line screen. 73 IV.1 1FI and 13C NMR data for vidalenolone (13). 84 V.1 Identified compounds for the TIC chromatogram shown in Figure V.3. 101 111.1 111.2 V.2 Effects on the yields of oxylipin from culture fed with six precursors. 107 List of Abbreviations AA Arachidonic Acid CC Column Chromatography CD Circular Dichroism CIMS Chemical Ionization Mass Spectrometry COSY 1H-1H DEPT Distortion less Enhancement by Polarization Transfer DMSO Dimethylsulfoxide EC Effective Concentration El Electron Impact EtOAc Ethyl Acetate FAB Fast Atom Bombardment FT Fourier Transform FTIR Fourier Transformed Infrared Spectroscopy GI Growth Inhibition GC Gas Chromatography HETCOR Heteronuclear Correlation Spectroscopy HEPE Hydroxyeicosapentaenoic Acid HETE Hydroxyeicosatetraenoic Acid HMBC Heteronuclear Multiple-Bond Coherence Spectroscopy HMQC Heteronuclear Multiple-Quantum Coherence HPETE Hydroxyperoxyeicosapentaenoic Acid HPLC High Performance Liquid Chromatography HRCIMS High Resolution Chemical Ionization Mass Spectrometry IC Inhibitory Concentration Chemical Shift Correlation Spectroscopy IPA Isopropyl Alcohol IR Infrared Spectroscopy LD Lethal Dose LT Leukotriene LX Lipoxin LO Lipoxygenase MS Mass Spectrometry NCI National Cancer Institute NMR Nuclear Magnetic Resonance NOE Nuclear Overhauser Effect NOESY Nuclear Overhauser Exchange Spectrometry PG Prostaglandin TGI Total Growth Inhibition TLC Thin Layer Chromatography TMS Tetramethylsilane TMSi Trimethylsilyl TOCSY Total Correlation Spectoscopy UV Ultraviolet Spectroscopy XHCORR Heteronuclear Chemical Shift Correlation Spectoscopy BIOACTIVE SECONDARY METABOLITES FROM MARINE ALGAE AND STUDY OF OXYDIZED ANANDAMIDE DERIVATIVES CHAPTER I : GENERAL INTRODUCTION Natural products have been a valuable source of medicine for centuries. Especially, natural products from terrestrial higher plants have been used extensively for practical pharmaceuticals from the 19th century until the late 1940s. A recent literature review summarized that about 120 drugs have been discovered from plants.' Remarkable examples include compounds with anesthetic (cocaine), skeletal muscle relaxant (tubocurarine), antimalarial (Cinchona alkaloids), antigout (colchicine), cardiac glycoside (Digitalis glycoside), and anticancer activity (taxol, vincristine, and podophyllotoxin). Since the collection of terrestrial samples is so heavily carried out, the study of natural products from this area has been extensively explored. Thus, it is highly difficult to discover novel compounds from terrestrial sources. On the other hand, the sea has been relatively unexplored. Since approximately three quarters of the earth's surface is covered by oceans, containing 1.8 million different species, marine species therefore exhibit a great diversity of life forms.' These different species have evolved over millions of years in a stable environment and natural selection has allowed the development of different chemicals as unique defense weapons. 2 Since the pioneering study of Bergmann on antibacterial agents from marine organisms in 1949,3 marine natural products have emerged as a valuable source of structurally novel compounds which may evolve to be potential drugs. Marine natural products research in the 1950s focussed on the study of organisms easily accessible from intertidal and shallow subtidal environments.' In the 1960s, the introduction of SCUBA (self-contained underwater breathing apparatus) allowed researchers to investigate organisms living at a variety of depths and locations. Today, an even deeper range of collections is achieved, to nearly 1000 m, by using manned submersibles (such as that designed by the Harbor Branch Oceanographic Institution (Fort Pierce, FL)). Clearly, collection of specimens at different depths increases the chance to discover potential new drugs . Prior to 1970, most research on marine natural products was limited basically to structural studies which included simple compounds such as mono- and sesquiterpenoids. From the late 1970s, considerable collaborative effort between marine chemists and pharmacologists has opened up biomedical applications for marine natural products targeting various neurotoxins, anticancer agents, antiviral agents, tumor promoters, and antiinflamatory agents. In the 1990s, due to the major development and swift growth of molecular biology, biochemical pharmacology, and genomic analysis, the research has become much more advanced and a new trend in drug discovery 3 programs has introduced the utilization of receptor or mechanism-based assays that evaluate very specific and selective activity:1'5 Chapter I of this thesis will provide a brief overview of selected fields including marine microorganisms, and natural products from cyanobacteria, followed by oxylipin chemistry. Chapters II, Ill, and IV respectively will describe in detail the discovery of secondary metabolites isolated from the marine cyanobacteria, Lyngbya majuscula and an Indonesian red alga, Vidalia sp. The following chapter V will focus on the quantitative analysis of oxylipin products from gametophyte cell cultures of a marine brown alga Laminaria saccharina. Chapter VI will concern semisynthesis of oxidized anandamide derivatives, new ligands for the cannabinoid receptor. Marine Microorganisms Microorganisms have had a significant influence on the development of therapeutic science since the finding that they not only cause infections, but also biosynthesize metabolites that can heal infections.4 Since the discovery of penicillin in 1929, microbial drug discovery focused on soil-derived bacteria and fungi has been the subject of intensive study that has led to the isolation of approximately 50,000 natural products.6 More than 10,000 of them are shown to have biological activity and over 100 microbial products are approved for use today as chemotherapeutic agents in the treatment of human and animal diseases. 4 Due to recent major advancements in screening methods and molecular biology, the study of microorganisms is recognized as a notable resource for drug discovery. Examples are the discovery of the bacterial metabolite FK-506 (1), an important immunosuppressant agent, and the fungal metabolite lovastatin (2), a compound for the treatment of high serum cholesterol. Although soil derived microorganisms have provided a fruitful source for many antibiotics and other bioactive metabolites, investigation of analogous microorganisms found in the marine environment has not been explored. This is mainly due to the lack of taxonomic data and difficulty in the isolation and culture of marine bacteria and fungi. Since marine sediments resemble soil, there must be huge diversity of bacteria present. In addition, the surfaces and internal spaces of marine plants and invertebrates provide microorganisms with unique habitats which are rich in nutrients and organic substrates.6 1 OCH3 2 5 Early studies on marine bacteria indicated the production of antimicrobial agents.' The first effort to elucidate marine bacterial metabolites revealed a highly brominated pyrrole antibiotic (3) isolated from the genus Alteromonas by Burkholder and co-workers. This seawater-derived bacterial compound was shown to have a high in vitro antibiotic activity against Gram-positive bacteria with minimum inhibitory concentrations (MICs = 6.3 200 ng/mL). However, further investigation showed that the compound (3) was not active in in vivo assays. Other studies revealed that it possessed some antitumor activity.8 Oncorhycolide (4), a structurally unique lactone, was produced by a seawaterderived bacterium obtained from a specimen taken from around a Chinook salmon (Oncorhyncus tshawytscha) net-pen farm.9 4 A pioneering and comprehensive study of marine bacteria has been initiated by the Fenical group at the Scripps Institution of Oceanography. His group has explored the isolation and cultural requirements of marine bacteria. Intensive efforts to obtain chemically novel bioactive compounds from a deepseawater bacteria have led to the isolation of a novel class of cytotoxic and antiviral macrolides, the macrolactins A (5) and B (6).b0 This Gram-positive 6 bacterium, which could not be identified by any taxonomic methods, grew slowly in a salt-based medium. The aglycon, macrolactin A (5), inhibits B16 F10 murine melanoma cancer cells in in vitro assay (IC50 = 3.5 ug/mL) and shows strong inhibition of mammalian Herpes simplex viruses (types I and II) (IC50 = 5.0 gg/mL) as well as human immunodeficiency virus (HIV) (IC50 = 10 gg/mL). CH3 5 CH3 6 Chemical investigation of a cultured marine-derived actinomycete of the genus Streptomyces, collected from the surface of a Sea of Cortez gorgonian octocoral Pacifigorgia sp., resulted in the isolation of the novel metabolites octalactins A (7) and B (8)." Octalactin A showed cytotoxicity against murine and human cancer cell lines (IC50 = 7.2 ng/mL, 500 ng/mL, respectively). Octalactin B is devoid of cytotoxic activity, suggesting that the epoxy functionality is necessary for this activity. In a continuing investigation, 7 0 0 H3C"ss' 0 ' CH3 ''' H3C"'s 'o QH CH3 CH3 CH3 8 7 H `CH3 %,, H 0 (3)\HH3ce Ws' N H K., OH CH3 CH3 H3C4:-­ H CH3 10 the Fenical group reported work with an actinomycete of the genus Streptomyces isolated from the surface of the jellyfish Cassiopeia xamachana. This surface associated bacterium produces in culture the complex bicyclic depsipeptides, salinamides A (9) and B (10).12 These compounds possess antibiotic properties with selective inhibition against Gram-positive bacteria. 8 However, the study of marine bacteria has given rise to another issue. Microorganisms involved in specific symbiotic relationships with macroorganisms may be responsible for the origin of some of the bioactive metabolites that have been isolated from such organisms. Several studies have demonstrated that some well-known marine toxins are in fact produced by microorganisms. The potent neurotoxin, tetradotoxin (TTX, 11) , has been known for many years to cause cases of fatal food poisoning around the world and was believed to be produced by pufferfish of the family Tetraodonitidae. A recent study revealed that TTX is truly produced by various unicellular marine bacteria.'15 Similarly, saxitoxin (12) and its derivatives, the well-known causative toxin of paralytic shellfish poisoning (PSP), have been known to be produced by several dinoflagellates.16 Although this fact has been generally accepted, another study reported that the true producer of 12 is a marine bacterium.' Since the discovery that 11 and 12 block the nerve membrane sodium channel, both toxins have become valuable tools for the study of neurophysiology and neuropharmacology. + NH2 HOH 12 9 Bioactive Metabolites from Cyanobacteria Cyanobacteria serve as an example of an evolutionary stage in the progress from primitive prokaryotes to eukaryotes. Cyanobacteria are similar to bacteria in their lack of organized nuclei and in their way of cell division, but are closer to higher plants in their ability to carry out oxygenic photosynthesis.' Members of the division Cyanobacteria appear ubiquitously, including freshwater, ocean, damp soil, pond, polar icecap, and even hot spring habitats. They are individually microscopic but some species aggregate to form macroscopic colonies. It has long been recognized that cyanobacteria produce toxins. Francis in 1878 reported in Nature the first intellectual delineation of the potential poisons produced by cyanobacteria, and livestock that drink at the surface of the water contaminated by cyanobacteria may become ill and even die.' Freshwater cyanobacteria produce numerous toxic metabolites. Microcystis aeruginosa, which belongs to the order Chroococcales, is a more primitive species than filamentous cyanobacteria such as Lyngbya majuscule. Study of blooms of this alga in eutrophic lakes have led to the characterization of microcystin (13)20 which was later designated microcystin-LR.21 This causative agent, a cyclic heptapeptide, is a powerful hepatotoxin which causes dose dependent chronic liver injury resulting in death in animals,22 and severe liver damage in humans.' 10 13 HN NH2 CH3 14 HN NH2 Extraction of the freshwater cyanobacterium Nodularia spumigena collected in the Baltic Sea and in New Zealand,' gave nodularin (14), a microcystin-related toxin which has turned out to be a liver carcinogen.25 Hapalosin (15), a novel cyclic depsipeptide, was isolated from the terrestrial cyanobacterium Hapalosiphon welwitschii.26 This metabolite was found to be a lead compound for a new class of potential anti-multidrug resistance (MDR) agents. Probably one of the most important discoveries of bioactive marine natural products is cryptophycin A (16).27 Cryptophycins are one of the largest 11 class of cyanobacterial secondary metabolites with 38 members. Cryptophycin A, the most active compound, was originally reported with gross structure as an antifungal agent by a group at Merck. 16 However, the Merck group decided not to study this compound further due to its extreme toxicity, an undesirable property for use as an antifungal agent. Cryptophycins were later revealed to be powerful antitumor agents exhibiting exceptional activity against a broad spectrum of solid tumors when inoculated into mice.28 Recent studies have demonstrated that cryptophycins are antimitotic agents which depolymerize microtubules and interact with the Vinca alkaloid binding domain of tubulin.29 12 On the other hand, marine cyanobacteria have also proved to be the source of many important bioactive secondary metabolites. The highly proinflammatory prenylated cyclic dipeptide, lyngbyatoxin A (17), was isolated from toxic strains of Lynbya majuscula collected in Hawaii and shown to be the causative agent of swimmer's itch.' 18 Majusculamide C (18), a novel cyclic nonadepsipeptide, was isolated from a deep water variety of Lynbya majuscula found in the Marshall Islands.' This peptide is a potent cytotoxin and showed a broad spectrum of antifungal activity toward plant pathogens. A Caribbean cyanobacterium, Hormothamnion enteromorphoides, is a producer of hormothamnin A (19), another cyclic 13 undecapeptide with moderate ichthyotoxic activity (LD50 - 5 j.tg/mL to the goldfish Carassius carassius).32 Notably, this peptide is closely related to the laxaphycin A (20) , an antifungal and cytotoxic cyclic peptide, isolated from Anabaena laxa (Nostocaceae), in both structural and biological features.33 19 20 Oxylipins from Marine Algae During the past three decades, many classes of marine organisms have produced eicosanoid-type natural products. The eicosanoids, a term referring to C-20 polyunsaturated fatty acids, are derived biosynthetically from the three most generally occurring C-20 polyunsaturated fatty acids found in invertebrates, vertebrates, and plants: arachidonic (eicosatetraenoic, 20:3 co6), 14 dihomo-gamma-linolenic (eicosatrienoic, 20:4 w6), or eicosapentanoic acid (20:5 w3).34 The eicosanoids play important roles in fundamental physiological regulation at the cellular level, and are particularly involved in various pathophysiological responses in mammals. For example, prostaglandins influence renal salt and water excretion by alterations in renal blood flow and by direct effects on renal tubules.35 They also participate in neoplastic diseases, regulate cellular and humoral immunity, as well as contract or relax many smooth muscles beside those of the vasculature.36 Leukotrienes cause hypotention, and contraction of most smooth muscles.' The major fatty acid for eicosanoid formation in mammalian cells is arachidonic acid which is released by the action of phospholipase A2.38 Arachidonic acid is metabolized through one of three notable pathways: the prostaglandin H synthase (cyclooxygenase) pathway, the lipoxygenase pathway or the cytochrome P450. This leads to three groups of metabolites, the cyclooxygenase products (prostaglandins and thromboxanes), the lipoxygenase products (hydroxyperoxy and hydroxyeicosatetraenoic acid, leukotrienes, and lipoxins), and cytochrome P450 products (hydroxyeicosanoids). Figure (1.1) presents an overview of the biosynthesis of the arachidonic acid cascade. Weinheimer and Spraggins in 1969 reported the pioneering discovery of the eicosanoid, 15-R-prostaglandin A2 (21), from the gorgonian Plexaura 15 Membrane Phospholipids Phospholipase A2 N., COOH Arachidonic Acid PGH Synthase 5- 12- Lipoxygenase Lipoxygenase PGH2 / \ Prostaglandins 5-HPETE 12-HPETE HHT's Leukotrienes Hepoxilins P450 Epoxide 15-HPETE /\ 15-HETE Thromboxane Figure 1.1 Cytochrome 15- Lipoxygenase Leukotrienes A HETE's diHETE's diHETE's Lipoxins The Arachidonic Acid Cascade. PG = Prostaglandins; HHT = Hydroxyheptadecatrienoic acid; HPETE/HETE = Hydroperoxy/Hydroxyeicosatetranoic acid diHETE = dihydroxyeicosatetranoic acid; Biosynthesis of the products of arachidonic acid in mammals. PG = Prostaglandins; HHT = Hydroxyheptadecatrienoic acid; HPETE/HETE = Hydroperoxy/Hydroxyeicosatetranoic acid; diHETE = dihydroxyeicosatetranoic acid Figure 1.1 homomalla.' Subsequently, more prostaglandins4"1 were isolated from P. homomalla and from a soft coral, respectively. Since the success in finding eicosanoids and prostanoids in corals, significant efforts were undertaken to explore the discovery of this class of chemistry in other marine organisms. As a result, a large number of oxidized metabolites of arachidonate and eicosanoate, 16 along with fatty acids of various chain lengths (e. g., C18, C20, C22) have been isolated from marine organisms. In order to cumulatively describe all of the fatty acid metabolites above, the new term "oxylipin" has been created for including all chain lengths of oxidized eicosanoid-related compounds.42 Since the first discovery of oxylipins from algae, namely PGE2 (22) and PGF2c, (23)43, from the red alga Gracilaria lichenoides, the marine algae have continued to yield an impressive array of oxylipins. Much of the pioneering work on marine oxylipins has been carried out by Gerwick and co-workers. The diversity and novelty of this class of chemistry isolated from marine organisms have been comprehensively reviewed by Gerwick.44-48 COON 21 OH COON 22 OH Relatively little has been reported on the production of oxylipins from cyanobacteria. The first oxylipin to be obtained from a cyanobacterium was the oxygenated fatty acid metabolite (24) from Lyngbya majuscule (Cardellina et al.).49 This compound (24) contains the C14 methoxytetradecenoic acid which 17 has been a common lipophilic constituent of many secondary metabolites found in marine cyanobacteria from the Pacific and Caribbean oceans. Similarly, malyngic acid (25), 9(S), 12(R), 13(S)-trihydroxyoctadeca-10(E), 15(2)-dienoic acid, was isolated from L. majuscule.' The structure of malyngic acid (25) was determined based on chemical and spectral data for the natural product and by its conversion to trihydroxystearic acid and degradation to deoxy-D-ribitol. QCH3 COOH 24 OH 25 The red algae (Rhodophyta) appear to be a prolific source of numerous unusual oxylipins. Over the last decade, the Gerwick group has demonstrated the ability of these algae to produce novel oxylipins. Two cyclopropyl and lactone containing oxylipins, constanolactones A (26) and B (27), were isolated from a red alga Constantinea simplex.' The absolute stereochemistries of these metabolites were determined by chiral separation of degraded fragments, CD analysis of the separately formed mono and dibenzoate derivatives and 1H NMR studies. 18 HO""".. HO"" 27 26 Another three new oxylipins (28-30), structurally similar dihydroxy oxylipins, were discovered from an Oregon coast red alga Farlowia moffis.' COON COOH OH 28 OH OH 29 30 Study on a Washington red alga Rhodymenia pertusa has led to the isolation of two major metabolites, dihydroxy-eicosanoic acid (31) and dihydroxy-eicosapentaenoic acid (32).46 The absolute stereochemistry of this diHETE and diHEPE was established by optical comparison to the known four diastereomers produced by synthesis.53 (-) Jasmonic acid (33) and 7+)­ isojasmonic acid (34) were discovered from a Black Sea red alga 19 COOH COOH 32 COON 0 34 Gelidium latifolium. In plants, these compounds play an important role as phytohormones, elicitors, and signal transducers.' They are biosynthesized through several enzymatic transformations. The green algae (Chlorophyta) have also produced some oxylipins but are generally less rich in C20 fatty acids. Two oxylipins, 12R-hydroxy-9(Z), 13(E), 15(Z)-octadecatrienoic acid (35) and 9(S)-hydroxy-10(E), 12(Z), 15(2)­ octadecatrienoic acid (36), were isolated from the freshwater unicellular green algae Duna liella COOH COON He 35 36 Two C10 metabolites (37-38) and five other C18 oxylipins (39-43) were obtained from an Oregon marine green algae Acrosiphonia coalita.56 The EZE­ 20 conjugated triene (38) was found to rapidly convert to the EEE-conjugated triene (37) under UV-irradiation. Observing this fact led to the hypothesis that the EZE-conjugated triene (38) is the true natural product and metabolite (37) is an artifact produced during isolation. COOH COOH COOH It was not until 7 years ago that brown algae (Phaeophyta) were found to be prolific producers of this class of chemistry. Examination of the temperate brown alga Cymathere triplicate collected from the Pacific Northwest revealed the C18 metabolite, cymathere ether A (44), and C20 metabolite, cymathere ether B (45).57 C. triplicate is an intertidal edible kelp in Washington and has an unique cucumber odor which arises from 2E-nonenal and 2E, 6Z-nonadienal, 21 both produced in cucumbers by the sequential actions of lipoxygenase and hydroperoxide lyase on fatty acids.' The absolute stereochemistries of these metabolites were predicted on the basis of biogenetic rational, CD analysis of w the p-bromobenzoate derivative and co-isolation of the known oxylipins, 12S­ HETE and 10S -NOTE. 00 HOliii....­ COOH 44 HOmi....­ COON COON Extracts of another temperate brown alga, Egregia menziesii, collected at Seal Rock State Beach, Oregon contained three new chlorinated and carbocyclic oxylipins, egregiachlorides A-C (46-48).59 A biogenetic pathway of 22 egregiachlorides A (46) was proposed where a 13-lipoxygenase introduces a hydoperoxide into the C18 precursor followed by the formation of an epoxy allylic carbocation. This intermediate cyclized to form a new cyclopentyl cation which was attacked by chloride anion at w3 carbocation to yield egregiachlorides A (46). 23 CHAPTER II. CURACINS B AND C, NEW ANTIMITOTIC NATURAL PRODUCTS FROM THE MARINE CYANOBACTERIUM LYNGBYA MAJUSCULA ABSTRACT Two new and potent antimitotic metabolites, curacins B and C, were isolated from a Curacao collection of Lyngbya majuscula. In addition, four curacin A analogs were prepared by semisynthetic efforts. The structures of the new curacins, along with the curacin A analogs, were determined by detailed spectroscopic analysis in comparison with curacin A. The absolute stereochemical configurations of curacins B and C were deduced to be 2R, 13R, 19R, 21S from interconversion with curacin A via a thermal rearrangement. The biological and biochemical properties of the new natural products and synthetic derivatives of curacin A were examined. 24 INTRODUCTION Different isolates of the marine cyanobacterium Lyngbya majuscula produce an amazingly wide array of biologically-active secondary metabolites; in this respect, reminescent of the rich biosynthetic capacity of some species of Streptomyces. The potent tumor promoters aplysiatoxin (1) and debromoaplysiatoxin (2),60 the antifungal lipopeptides majusculamides A (3) and B (4),61 and the structually unusual pyrrolic compounds, pukeleimides A-F (5-10)62'63 are examples of the range of metabolites isolated from this organism (Figure 11.1). Our survey for the discovery of bioactive secondary metabolites from marine algae has focussed on a Curacao (Caribbean) collection of Lyngbya majuscula which has resulted in the isolation of several novel compounds. Antillatoxin (11) is a powerful ichthyotoxic cyclicdepsipeptide from L. majuscula collected at Curacao (Caribbean).64 The absolute stereochemistry of 11 was determined on the basis of a combination of analysis by chiral phase TLC, molecular modeling, NMR and CD spectroscopy. An extract of L. majuscula from Playa Kalki, Curacao was the source of kalkitoxin (12), a thiazoline containing lipid.65 This compound (12) shows potent ichthyotoxicity (LC50 = 0.05 lig/mL to Carassius auratus). During the past five years, a major effort in our laboratory has been the isolation, structure elucidation, and biological evaluation of curacin A (13).66,67 25 CH3 CH3 o;;;-------o OH 0 4cH3 T T Q CH3 8H3 0 OH 1. R= Br OH 2. R=H H3C CH3 Figure II. 1 Structures of selected secondary metabolites from Lyngbya majuscula. 26 H3C-N H H i CH3 CH3 0 CH3 CH3 0 12 In initial screening at Syntex, a crude extract of L. majuscula was highly cytotoxic against a monkey Vero cell line. Following evaluation of this extract in the National Cancer Institute (NCI) in vitro 60-cell line screen which showed a potent antiproliferative and cytotoxic activity (selective for colon, renal and breast cancer-related cell lines), curacin A was isolated as the major bioactive species. Further study has revealed curacin A's ability to inhibit tubulin polymerization, inhibit colchicine binding to tubulin, and inhibit cell proliferation. 67 In continuing investigations of this organism for minor metabolites of related structure, we have isolated and characterized two new natural products, curacins B (14) and C (15).68 Both of these are toxic to brine shrimp, demonstrate strong cytotoxicity against murine L1210 leukemia and human CA46 Burkitt lymphoma cell lines, and in the National Cancer Institute in vitro 27 60-cell line assay (Figure 11.2), show a potent antiproliferative activity to many cancer-derived cell lines in a manner characteristic of antimitotic agents. RESULTS AND DISCUSSION Isopropanol preserved Lyngbya majuscula collected from Curacao was extracted (CHCI3 /Me0H, 2:1) and chromatographed over silica gel to give a mixture of curacins A-C (13-15) contaminated by fatty acids and phytol (Figure 11.3). These contaminents were removed by methylation (CH2N2) and acetylation (Ac20/pyridine) of the curacin-containing fraction followed by NP-HPLC to yield two peaks. The first peak was curacin A (13) (10.3%) while the second was a mixture of two new curacins by1H NMR spectroscopy and GC-EIMS analysis. Repeated RP-HPLC (C-8) of this latter fraction eventually gave pure curacin B (14) and C (15) as 4.7 % and 0.3 % of the extract, respectively. Compounds 14 and 15, both analyzing for C23H35NOS by HRFABMS (positive ion, [M+H]+), displayed 1H NMR and 13C NMR bands indicative of four 17 CH3 H2 9 OCH3 S 7 13 OCH3 22 CH3 18 H 20 CH3 OCH3 HC 15 CH3 co lethality). 50% for required concentration LC50, inhibition; growth total for required concentration TGI, inhibition; growth 50% for required concentration (G150, screen vitro in line cell 60 Institute Cancer National the from obtained mixture (95:5) C and B curacins of graphs Mean 11.2 Figure 1 4 1 1 1 .3 1 I 2 I .1 41 .2 41 1 .3 I I 4 la 343 ^ La IN 1 1 I 4 1 31 .1 43 J 4 1 4 1 .1 .2 .3 I Doh MD MO 4.23 400 020 7.11 55V .---­ .. - --..--­ --------- ....... ...-....-.--...­ < AIDA. 4017 > a a -741 .140 440 111.349 44 44 4.0 434 1.470 144411431 on 420 0 400 4 CM 143 4 MCP7MDR.R1111 a 40 .31 .73 100 40/14410231/A1CC a 410 4.00 0 ---.....-..... Caw Sam ...........-......-----.....-......-.......-...--­ -...­ ........ -- ..... ---...-­ ----...-- ............... ­ ..... .,....- 1 4.11 14CP7 441 4.74 131.1443 o 4.00 ICJ ------- -- ..... .----­ -...----------­ -....---­ .............. ....-­ -.----.-----­ Cao 10501 44D a 4.0 R30.451 4.40 4.7 .441 420 23 JO NNE 447 Ma 420 ACM 40 CAK1.1 412 JIM 4. 1 4.33 473 81112C owEmomor 4.113 4.0 403 0 7.00 144.31 11440 a 403 400 MI 4.41 7364 - ------.­ a----- - .. ----- -------- Cam hal o 4431 OVCA1741 4.4 AM 4.00 4.00 i 41.03 OVC/414 , 443 ........ ..--.--------. .-­ --..---,---­ - --­ --.-.--­ UACC.41 114144.4 4.30 443 nom 4.03 -7.20 1.1ACC.4.17 4.44 sc-mstAs 4.0) 4. 403 4.0) s 000 0Moneerml -.--...--....­ 1OROVI 442 400 4.40 4.14 OVCARol 40 a 4.31 OVCAR4 0 0 0 40 Ea 443 4401.3 am 4/73 NI 4.2 4.1.0114 4 4.00 4 RAVI 1.32 420 420 4131 ) 4.1 <La 4.11 104.3411L4 41 ....-..-.-... --- ...... .4...o.. - - ....... - . - - Mama. 40) a .0 320 0 0 4.14 4.05 imm. gm 443 470.11 44 411143 7.51 4.11 1/231 . 402 401 a 40 --- - .. ...... . -...­ Caw CRS ­ -. -­ ..... 1014 .741 403 4.03 40.393 s 4 1444 a 410 400 41 443 -7.73 403 < 4.00 .7.13 NCI, 1 III RCT4 1.311 wrn .7.34 .7.11 14M12 .4 Mali 4.1 1., 443 1404901 203 COLO -...­ -...­ ......--..­ ....- Cana han AZ .7 4.00 400 40 NCI4423 4.0 4.15 141441304 4.01 403 ma 14141400 434 4. < 14141122 110043 110044 443 403 4.10 41 4.00 a 400 ... - .. - -- .. .­ war 401 - ­ ---..--- .­ -------- -. ­ - ...... --­ .- -­ ..... ­ - ­ 440 . . 420 C Las Cen Madan L MOM= .4.0) 4.13 11124V14 4.00 4.00 .. .711 410 CO 4.44 14041120 4.01 4.00 4.30 4102 111.40070 > OZR4C114 403 403 4.00 4.40 420 410 441 4.00 403 4.03 MOLT. 440 401 0011414301 La* LCD LC-14 Lei. TOI 1,4.701 OM OM Log. Una hal/Con 29 Lyngbya majuscula (Curacao collection) 543 mgs of 2.39 gms total Vacuum Chromatography 5% E/H 5%110%110%115%1201:Y0 30%150%175%175%100°AI ...._.._., 314 2 mgs Vacuum Chromatography 1% E/H 1c/011.5cY 1 1.5%1 2% 13% 14% 1 5°% 0110°% 01100cYcl i ca. 200 mgs 1 158 mgs CH2N2 1 HPLC 4% EtOAc /Hexanes minor Curacin B & C (29 mgs) major Curacin A (44.2 mgs) Ac20/Pyr. HPLC, C-8 column 64% CH3CN/H20 Curacin B 20 mgs Curacin C 1.1 mgs Figure II. 3 Flow chart of fractionation of Lyngbya majuscula H-8 H-9 11-7 Curacin A 144t, I Curacin B 5'5 6.0 5.5 5.0 4.t 5 4.0 3.5 3.0 25 n Figure 114 Comparison of 1F1 NMR spectra of curacins A, B, and C. Table 11.1 11-1 and 13C NMR data for curacins A (13), B (14), and C (15) Curacin A Curacin B Curacin C Position 1H(Jin Hz)a la 2.79 (dd, 10.7, 10.0) 3.09 (dd, 10.7, 10.0) 5.11 (m) 5.69 (dd, 10.7, 8.7) 5.45 (bdt, 10.7, 7.1) 2.11 (m) 2.11 (m) 5.58 (bdt, 15.0, 7.0) 6.38 (dd, 15.0, 10.8) 6.02 (d, 10.8) b 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20a 2.23 (m) 1.70 (m) 3.09 (m) 2.26 (m) 5.87 (m) 5.07 (m) 1.70 (s) 13cb 39.96 74.31 131.32 130.84 28.14 33. 12 131.32 127.86 125.50 136.40 35.77 32.15 79.91 38.02 135.32 116.80 16.56 1H(Jin Hz)a 2.79 (dd, 10.7, 10.0) 3.09 (dd, 10.7, 8.4) 5.12 (m) 5.69 (bdd, 10.9, 7.3, 0.9) 5.45 (bdt, 10.9, 7.3, 0.9) 2.11 (m) 2.26 (m) 5.40 (dt, 9.4, 7.4) 6.35 (m) 6.35 (m) 2.23 (m) 1.69 (m) 3.09 (m) 2.26 (m) 5.87 (m) 5.07 (m) 1.69 (s) 168.36 1.65 (m) 0.75 (td, 8.2, 4.2) 1.20 (m) 21 0.98 (m) 22 1.21 (d, 6.3) 2') (OMe) 3.18 (s) 0.75 (td, 8.1, 4.4) 15.98 12.39 56.31 1.21 (m) 0.99 (m) 1.22 (d, 6.3) 3.19 (s) a300 MHz in C6D6, b75 MHz in C6D6 1H (J in Hz)a 13Cb 39.99 2.80 (dd, 10.5, 10.1) 3.09 (dd, 10.5, 8.6) 5.11 (m) 5.69 (dd, 10.5, 8.9) 5.45 (m) 2.11 (m) 2.11 (m) 5.57 (bdt, 15.0, 6.6) 8.51 (dd, 15.0, 10.8) 5.97 (d, 10.8) 39.94 74.33 131.40 130.91 28.15 27.79 128.83 125.84 120.74 138.73 36.26 32.26 79.95 38.06 135.32 116.84 16.39 1.65 (m) 20.13 14.24 15.99 12.35 56.37 74.31 131.30 1.70 (m) 130.85 28.16 33.12 131.30 127.67 126.42 136.62 28.16 32.16 79.70 37.85 135.23 116.84 23.65 169.34 20.11 0.75 (td, 8.1, 4.1) 14.21 2.38 (m) 1.62 (m) 3.09 (m) 2.24 (m) 5.85 (m) 5.07 (m) 1.74 (s) 168.42 19.87 13.82 b 13cb 1.23 (m) 0.98 (m) 1.22 (d, 6.1) 3.20 (s) 15.98 12.31 56.23 32 bonds and one C=X functionality. Therefore, in consideration of the molecular formula, both contained two rings in similarity to curacin A (13). Comparison of the 1H and 13C NMR features of the two new compounds with that of curacin A revealed a close comparability of all three metabolites (Figure 11.4 and Table II. 1). The only NMR regions of significant difference between them was in that assigned to the chemical shifts and coupling patterns for the C-8 to C-11 diene and the associated C-17 methyl group. Hence, it was concluded that curacins B (14) and C (15) represented geometrical isomers of curacin A in this diene region of the molecule. In our prior work on curacin A (13),66 the geometry of the C-8 to C-11 diene was established as 7(E), 9(E) on the basis of a 15 Hz coupling between H-7 and H-8 (Table II. 1) and a strong nOe interaction between the C-17 methyl group and H-8. Correspondingly, in curacin B (14), a Z geometry for the C-7 olefin was deduced by observing a 9.4 Hz coupling between H-7 and H-8, an E geometry for the C-9 olefin by observing the upfield 13C chemical shift for C-17 methyl (16.4 ppm)69 and an nOe between C-17 methyl and H-8 (Figure 11.5). In curacin C (15), the combination of a 15.0 Hz coupling between H-7 and H-8 with 1H and downfield 13C nmr chemical shifts for C-17 methyl (23.6 ppm) indicated it was the 7(E), 9(Z) isomer. The absolute configuration of 14 was defined through thermal interconversion with 13,7° the absolute configuration of which we recently defined as 2R, 13R, 19R, 21S (Figure 11.6). Refluxing a 95:5 mixture of 14 and 15 in 2,2,4-trimethylpentane (99° C) for 3 h yielded a 2:1:1 mixture of all three 33 Figure IL5 Geometrical configuration of conjugated diene of curacin B (14) determined by NOESY in CDCI3. 34 curacins (13-15) by GC-EIMS (Figure 11.7). It is intriguing that all curacins have unusual peak shapes in GC. Although there is no clear explanation for this, it is likely that the unstable cis cyclopropyl moiety of the structure might undergo thermal rearrangment during the GC-MS process. This prediction is based on the experimental observation where 19-epi-curacin A (19) possessing the trans cyclopropyl ring showed a single peak in GC. The compound 13 produced in this experiment was purified by HPLC and had identical spectral properties to authentic 13 (1H-NMR, GC-EIMS, UV) and a comparable optical rotation ([425 +77°, c = 0.1; lit. [425 +86°, c= 0.64, CHCI3). Therefore, curacin B (14) possesses the same 2R, 13R, 19R, 21S stereoconfiguration as curacin A (13). / OCH3 OCH3 Curacin A (13) Isooctane, reflux (99 °C) OCH3 CH3 CH3 Curacin B (14) \ H3C z E Curacin C (15) Figure II. 6 Thermal isomerization of curacins A, B, and C Additionally, because HPLC-purified 14 and 15 had comparable optical rotations (see Experimental), it is likely that 15 also possesses 2R, 13R, 19R, 21 S absolute stereochemistry. CH3 q. n.ance TIC: 1. /0-5-2.0 200000 . mice TIC: 111-38.0 1000000 800000 150000 600000 1 100000 400000 50000 - 200000 8.00 10.00 12.00 14.00 0 16.00 18.00 ice -> curacins B and C (95:5), before refluxing a) A ......18. 0 Imo -> TIC: I/-51-1.0 kNiNeese 8.00 10.00 14.00 16.00 18.00 20.00 22.00 refluxing B and C in TMP for 1 h once 700000 12.00 TIC: III-38-2.0 600000 600000 500000 500000 400000 400000 300000 300000 200000 200000 C­ 100000 0 Time -> 8.00 c) 10.00 C­ 100000 0 12.00 14.00 16.00 18.00 refluxing B and C for 90 min. 20.00 ice -> 8.00 10.00 12.00 14.00 16.00 18.00 refluxing B and C for 2 h Figure 11.7 GC-MS traces for thermal isomeration of curacins A, B, and C. 20.00 36 Three experiments were conducted to confirm that 14 and 15 are true natural products of this cyanobacterium and not extraction/isolation artifacts. First, a cold extraction of the preserved cyanobacterium was shown by GC­ EIMS to contain the same proportions of 13, 14, and 15 as obtained from the above warm extraction. Second, direct heating of pure 13 in CH2Cl2 -MeOH (2:1) at 35° C for 10 h did not produce detectable quantities of 14 or 15. And third, pure 13 refluxed in aqueous CH2Cl2 -MeOH (2:1) for 90 min. also did not give 14 or 15. It should be noted that in these latter two experiments, 13 was heated at a temperature lower than that shown to affect isomerization of the 14/15 mixture into 13. In the National Cancer Institute (NCI) in vitro 60-cell line screen, a 95:5 mixture of curacins B and C showed potent antiproliferative activity to many cancer derived cell lines in a manner characteristic of antimitotic agents.68'71 Some structurally diverse compounds, most of which originated from natural products, have the ability to bind to tubulin or microtubules and inhibit cell proliferation by acting on spindle microtubules. These microtubule-active agents can be divided into two classes: those that inhibit the polymerization of spindle microtubules or depolymerize existing spindle microtubules and cause the loss of cellular microtubules, and those that stablize microtubule polymerization and cause the formation of intracellular microtubule bundles. Example of the former includes colchicine, the vinca alkaloids, rhizoxin,72 dolastatins,73 and curacins. The latter includes taxol, related taxanes and epothilones.74 Many antimitotic drugs bind to the colchicine (16)-binding 37 0 11 NHCCH3 H 140 N H SS le O H3COOC N 16 OCH3 17 CH3 OCOCH3 HO COOCH3 CH3 H2 OCH3 18 SCH3 OCH3 NH (:) 21 CH3 H2 H3C CH3 .........0 H2 S -S OCH3 0 NH HN CH3 H3C 0 OCH3 22 Figure II. 8 Natural antimitotic agents and synthetic derivatives of the curacins 38 domain and inhibit the binding of colchicine to tubulin while others bind to the vinblastin (17)-binding domain and inhibit the binding of vinblastine to tubulin (Figure 11.8). Most known colchicine-type tubulin inhibitors possess a common topographical similarity which provide a structure-activity relationship of that binding site. Curacin A is an example of the former case that inhibits tubulin assembly by binding to the colchicine site on tubulin.' This is intriguing because the structure of curacin A has little similarity to other drugs that bind to the colchicine-site. Consequently, in order to better understand how curacin A resembles these other drugs and to get further insight into the molecular mechanism of tubulin-ligand interaction at this site, some curacin A derivatives (18-22) were prepared.' Heat treatment of curacin A produced cleavage of the cyclopropyl ring to yield cyclopropyl ring opened (CPRO) curacin A (18). CPRO-curacin A was obtained as an optically active pale yellow oil which gave a molecular formula of C23H36ONS by HRFABMS. The 13C NMR and HMQC identified resonances for all 23 carbons. The 1H NMR and COSY showed features similar to that of curacin A except for the absence of cyclopropyl resonances in 18 and extra olefin signals at 8 4.96 (ddd, J . 10, 1.7, 1.1, Hz, H-22b), 8 5.02 (ddd, J . 17, 1.7, 1.7 Hz, H-22a), and 8 5.76 (m, H-21) (Figure 11. 9 and 10). Another curacin A derivative, 19-epi-curacin A (19) was produced accidentally by the treatment of curacin A with acetic anhydride and pyridine. In fact, the original intention of this reaction condition was to remove the undesired impurity, phytol, from the mixture of curacins B and C. j LI 6.0 5.5 5.0 4.5 4.G PPM 3.5 3.0 2.5 Figure 11.9 1H NMR spectrum of cyclopropyl ring opened curacin A (18) in C6D6 2.0 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5 6.0 6.0 5.5 5.0 4.5 4.0 3.5 3.0 2.5 PPM Figure 11.10 1H -1H COSY spectrum of cyclopropyl ring opened Curacin A (18) in C6D6 0 6.5 6.0 5.5 5.0 4.5 4.0 1 3.5 PPM 3.0 2.5 2.0 1.5 Figure 11.11 1F1 NMR spectrum of 19-epi curacin A (19) in C6D6. 1.0 6.0 5.5 5.0 4.5 4.0 3.5 3.0 2.5 2.0 1.5 PPM Figure 11.12 1H-1H COSY of 19-epi curacin A (19) in C6D6 1.0 6.5 6.0 5.5 5.0 4!5 4.0 3.5 PPM 3.0 2.5 2.0 1.5 1.0 Figure 11.13 11-1 NMR spectrum of thiazoline ring opened curacin A (20) in C6D6 10 11 ct) .5 .04 1I 140 160 180 7.0 6.5 I 6.0 /0. 5.5 5.0 4.5 4.0 PPM 3.5 3.0 2.5 4 2.0 1.5 1.0 1 Figure 11.14 HMBC plot of thiazoline ring opened curacin A (20) in C6D6 PPM 6.5 1 6.0 5.5 1 5.0 4.5 4.1 0 3.I 5 PPM 3.0 2.5 2.0 Figure 11.15 'H NMR spectrum of derivative 21 in C6D6 1.5 1.0 .A.2 Athdukl, 6.5 6.0 1 5.5 , 5.0 1 4.5 4.0 3.5 3.0 2.5 PPM Figure 11.16 11-I NMR spectrum of curadisulfide A (22) in C6D6. 2.0 1.5 47 Compound 19 was prepared as a pale yellow oil. The molecular ion [M + was observed at m/z 374, which is consistent with a molecular formula of C23H35ONS by HRFABMS. Its 11-1 NMR spectrum showed essentially the same signals as those present in curacin A. The observed differences between the 1H NMR spectrum of 13 and 19 were consistent with 19 being the C-19 epimer of curacin A, where only the methyl and methine resonances of the cyclopropyl ring were shifted upfield or downfield (e. g., H-19 at 6 1.65 to 6 1.55; H-20a at 8 0.75 to 8 0.43, H-20b at 6 1.20 to 8 1.34; H-21 at 6 0.98 to 8 1.49; H-22 at 6 1.21 to 8 0.85) (Figure 11.11 and Figure 11.12). Thiazoline ring opened (TRO) curacin A (20) was obtained from the treatment of curacin A with acetic anhydride and D20. TRO curacin A (20) was also obtained a pale yellow oil. This derivative exhibited a molecular formula of C25H40NO3S based on its HRFABMS data. The IR absorptions at 1678 and 1650 cm-1 were attributed to an amide or thio ester carbonyl group. The structure of compound 20 was identified by comparing its 1H and 2D NMR spectra with those of curacin A (Figure 11.13 and Figure 11.14). The main differences in the 11-1 NMR data of 20 and 13 were the presence in compound 20 of the methyl resonance (8 1.58) and the upfield shift of H19 (8 1.8), and the downfield shifts of H20a (8 0.65), H20b (8 1.11), and H22 (8 1.12). In addition, 13C NMR spectroscopy revealed the presence of amide carbonyl (8 167.99) and thio ester carbonyl (8 196.96) resonances in the derivative. 48 The cyclopropyl ring was further cleaved by treating 20 with methyl lithium in THF, followed by methyl iodide to give the methyl thio ether derivative of curacin A (21). Compound 21 was isolated as an oil that gave a molecular ion at m/z 365 in GC-EIMS corresponding to a molecular formula of C21H3502NS. Analysis of 1H- NMR data for 21 revealed the absence of cyclopropyl and thiazoline rings, and the presence of amide carbonyl and methyl thio group. These were further indicated by GC-EIMS by observing fragments at m/z 318 ([M - SCH3] +), 304 ([M - SC2H5] +), 233 ([M - C5F1,00NS] +). The 11-1 NMR spectrum showed signals at 8 1.55 (3H, s), 8 1.91 (3H, s), and 8 4.70 (1H, br) for the vinyl methyl, methyl thio, and amide protons, respectively (Figure 11.15). Curadisulfide A (22) was produced by treating 20 with sodium hydroxide in methanol. The FABMS of 22 gave a [M + Hr at m/z 701. A detailed mass spectrum revealed its dimeric nature by observing a fragment ion at m/z 305. The 1H NMR spectrum of 22 was very similar to that of 20, except for the absence of resonances for the methyl cyclopropyl moiety (Figure 11.16). Using a purified tubulin preparation, these derivatives were examined, as well as pure curacins A and B, for their effects on the inhibition of tubulin polymerization, and radiolabeled colchicine binding to tubulin, and cytotoxicity to a cancer cell line (Table 11.2).71 Curacin B exhibited strong inhibitory activity on purified tubulin assembly and it was half as inhibitory of colchicine binding to tubulin as that of curacin A. Curacin B was approximatly 10-fold less active in 49 Compound Inhibition tubulin Polymerization IC50 ± S.D. (pM) Inhibition colchicine Inhibition of MCF-7 Brine Binding Growth Shrimp `3/0 Inhibition IC50 ± S.D. (M) IC50 (nM) Curacin A 0.72 ± 0.2 94 ± 2 0.038 ± 0.01 8 Curacin B 0.82 ± 0.2 56 ± 2 0.32 ± 0.06 38 Curacin C 2.3± 1 10 ± 2 3.6 ± 0.8 54 18 0.92 ± 0.2 83 ± 6 0.30 ± 0.2 19 0.77 ± 0.2 88 ± 11 0.09 ± 0.01 20 1.5 ± 0.4 9±8 4.2 ± 2 >1000 21 3.3 ± 0.8 12 ± 15 5.2 ± 3 >1000 22 120 ± 20 8 Colchicine 1.4 15 700 1.7 ± 0.09 Podophyllotoxin 1.6 ± 0.03 83 1.7 ± 0.09 Table 11.2 Biological activities of curacins on cell growth, tubulin polimerization, colchicine binding, and brine shrimp toxicity. its inhibitory effects on the growth of MCF-7 breast cancer cells than curacin A. Curacin C turned out to be remarkably less active than curacin A in the tubulin­ based assay. In the inhibition of tubulin polymerization, it was almost 3-fold less active and showed only weak inhibitory capability for the binding of [3H]colchicine to tubulin. On the other hand, it had IC50 values almost 100­ timeshigher than that of curacin A to the growth of MCF-7 cancer cells. Based on these results, it is apparent that geometrical isomerization of the C-7 and C-9 olefins has a pronounced effect on the ability of these compounds to inhibit 50 tubulin polymerazation, to inhibit colchicine binding to tubulin, and to increase cytotoxicity. Cyclopropyl ring opened (CPRO) curacin A (18), unlike our expectation, was slightly less active than curacin A as an inhibitor of both tubulin polymerization and colchicine binding, but was 10-fold less cytotoxic than curacin A on MCF-7 cancer cells. The activities of 19-epi-curacin A (19) were basically identical to those of curacin A. Thiazoline ring opened (TRO) curacin A (20) still possessed good inhibitory properties of tubulin assembly and colchicine binding, but was about 9-fold less active than curacin A at inhibiting the growth of cancer cells.' Surprisingly, the methyl thio ether derivative of curacin A (21) retained good activity as an inhibitor of tubulin assembly as it was only 4-5 fold less potent than curacin A. However, compound 21 was also a very weak inhibitor of colchicine binding to tubulin and of cancer cell proliferation. Curadisulfide A (22) was the least active of the curacin A derivatives in the inhibition of tubulin polymerization. Nevertherless, it retained some activity as an inhibitor of both MCF-7 cell growth and colchicine binding. EXPERIMENTAL General Experimental Procedures. NMR spectra were recorded on Bruker AM 400 and AC 300 spectrometers in the solvent specified. Chemical shifts were referenced to solvent (C6D6 signal at 7.2 ppm for 1H NMR and 128 ppm for 130 NMR). Mass spectra were recorded on Kratos MS 50 TC and Finnigan 4023 mass spectrometers. Gas chromatography/mass spectrometry 51 (GC/MS) was carried out utilizing a Hewlett-Packard 5890 Series II GC connected to a Hewlett-Packard 5971 mass spectrometer. UV spectra were obtained on a Hewlett-Packard 8452 A UV-VIS spectrophotometer and IR spectra on a Nicolet 510 spectrophotometer. All solvents were distilled from glass prior to use. Collection, Extraction, and Isolation Lyngbya majuscula was collected on 13 December 1991 from CARMABI beach, Curacao and preserved in i-PrOH at low temperature until workup. Isopropanol preserved Lyngbya majuscula was extracted with CHC13/Me0H (2:1). Gradient vacuum liquid chromatography of the crude extract (543 mg) with EtOAc /hexanes gave a mixture of curacins contaminated by fatty acids and phytol. These fatty acids were treated with an excess of ethereal CH2N2 for 5 min and then concentrated in vacuo. Subsequently, the mixture was treated with 0.5 mL of acetic anhydride in pyridine (0.5 mL). The resulting mixture was then ready for HPLC. Isolation of Curacins B (1 4) and C (1 5). Repetitive HPLC on a Phenomenex Si column (10 mm, 500 x 10 mm) of the methylated mixture with 4% (v/v) EtOAc /hexanes resulted in curacin A (44.2 mg) and B (29 mg) containing C. The latter was subjected to further HPLC on a Phenomenex phenosphere C8 column (10 mm, 250 x 4.6 mm) with 64% CH3CN/H20 to afford curacin B (20 mg, 69%) and C (1.1 mg, 4%). Curacin B showed the following: [a]25D +620 (c = 0.84, CHCI3); UV Amax (MeOH) 242 nm (E. = 22000); IR vmax (film): 2975, 2930, 2874, 1615, 1441, 52 1379, 1093, 1075, 1056, 998, 967, 914 cm-1; LREIMS m/z (rel. int. °A)) 373 (3), 342 (9), 332 (13), 274 (8), 181 (28), 180 (100), 166 (16), 140 (14) 99 (17), 91 (27), 85 (20), 79 (31), 67 (17), 55 (15); HR FAB MS (positive ion) obs. [M + F1]-+' at m/z 374.2526 (0.9 mmu error for C23H36ONS); Curacin C showed the following: [a]25D +62° (c= 0.84, CHCI3); UV ,max (MeOH) 242 nm (E = 25000); IR 'Umax (film): 2970, 2927, 2885, 1617, 1438, 1095, 1074, 1054, 998, 964, 914 cm-1; LREIMS m/z (rel. int. %) 373 (4), 342 (9), 332 (12), 274 (8), 181 (29), 180 (100), 166 (16), 140 (14) 99 (17), 91 (26), 85 (25), 79 (30), 67 (18), 55 (16); HRFAB MS (positive ion) obs. [M + H]+ at m/z 374.2532 (1.5 mmu error for C23H36ONS); Thermal lsomerization of Curacin B to A and C. To 6 mg of a 95:5 mixture of curacins B and C was added 2.5 mL of trimethylpentane. The reaction mixture was refluxed for 3 hr and then concentrated in vacuo . The concentrate was diluted with 4% EtOAc /hexanes, filtered through on sintered glass, and subjected to HPLC (Phenomenex Maxil 10 µm Si, 500 x 10 mm, eluted in 4% (v/v) EtOAc /hexanes, flow rate at 9 ml/min, UV detection at 254 nm) yielding curacin A (0.9 mg, 15% yield) and a mixture of curacins B and C (0.8 mg, 13%) Confirmation That Curacins B and C are Natural Products. First, preserved Lyngbya majuscula was cold-extracted by grinding in a mortar with liquid N2. Solvent (CH2Cl2 -MeOH, 2:1, 30 mL) was added to the algal powder 53 and stirred for 1 h. The algal material was removed by filtration and the solvent reduced in vacuo, dissolved in minimal 4% EtOAc /hexanes, passed through a plug of Si gel to remove polar compounds, and analyzed by GC-EIMS. This gave a similar profile of curacins A-C to that from the hot extracted material. A second approach involved heating pure curacin A (2.5 mg) at 35° in CH2C12­ Me0H (2:1, 4 mL) for 10 h, followed by GC-EIMS analysis. Finally, curacin A (2 mg) was refluxed in CH2Cl2 -MeOH (2:1, 3 mL) containing 4 drops H2O for 90 min, and then analyzed by GC-EIMS. These latter two experiments showed only the presence of curacin A. Preparation of CPRO-curacin A (1 8). To 15 mg of curacin A (40 limo') was added 2.5 mL of toluene. The reaction mixture was heated at reflux for 4 hr. The resulting solvent was allowed to cool down to RT and removed in vacuo. The resulting residue was dissolved in 4% EtOAc /hexanes and filtered through the sintered glass. The filtrate was purified by HPLC (Phenomenex Maxil 10 Si, 10 mm, 500 x 100 i. d., 4% (v/v) EtOAc /hexanes, flow rate at 9 mL/min, UV detection at 254 nm) to afford CPRO-curacin A (18) (7.3% yield) which showed the following: [a]2210 +1100 (CHCI3, c = 0.1); UV Xmax (MeOH) 242 nm (e = 25000); IR Dmax: 2977, 2925, 2868, 1621,1437, 1096, 963, 913 cm ' ;IH NMR (300 MHz, C6D6) 8 6.38 (dd, 1H, J= 15, 10.8 Hz, H-8), 6.05 (d, 1H, J= 10.8 Hz, H-9), 5.87 (m, 1H, H-15), 5.76 (m, 1H, H-21), 5.62 (dd, 1H,J = 10.7, 8.4 Hz, H-3), 5.6 (dt, 1H, J= 15, 7 Hz, H-7), 5.46 (dt, 1H,J =10.7, 7.1 Hz, H-4), 5.14 (m, 1H, H-2), 5.08 (m, 2H, H-16), 5.02 (ddd, 1H,J = 17, 1.7, 1.7 Hz, H-22a), 4.96 54 (ddd, 1H,J = 10, 1.7, 1.1 Hz, H-22b), 3.18 (s, 3H, -OCH3), 3.09 (m, 1H, H-13) 3.09 (dd, 1H,J =10.7, 8.4 Hz, H-lb), 2.77 (dd, 1H, J . 10.7, 8.7 Hz, H-1a), 2.5 (m 2H, H-19), 2.42 (m, 2H, H-20), 2.25 (m, 2H, H-14), 2.25 (m, 2H, H-11), 2.15 (m, 2H, H-6), 2.15 (m, 2H, H-5), 1.70 (s, 3H, H-17), 1.65 (m, 2H, H-12); 13C NMR (75 MHz, Cal)) 8 168.68 (C18), 137.27 (C21), 136.50 (C10), 135.35 (C15), 131.32 (C7), 131.14 (C3), 130.78 (C4), 127.93 (C8), 125.53 (C9), 116.78 (C16), 115.39 (C22), 79.97 (C13), 74.53 (C2), 56.31(-0CH3), 39.83 (C1), 38.06 (C14), 35.80 (C11), 33.98 (C19), 33.13 (C6), 32.19 (C12), 31.62 (C20), 28.19(C5), 16.59 (C17); LREIMS m/z (rel. int. %) 373 (4), 332 (15), 181 (32), 180 (100), 140 (16) 119 (17), 105 (20), 93 (20), 91 (27), 85 (20), 79 (33), 67 (16), 55 (14); HRFABMS (positive) obs. [M + F1]-1- at m/z 374.2494 (2.4 mmu error for C23H36ONS) Preparation of 19-epi-curacin A (1 9). To 20 mg of curacin A was added 1 mL of pyridine and 1 mL of reagent grade acetic anhydride. The reaction mixture was stirred at rt overnight. The solvents were then removed in vacuo. The concentrate was diluted with 4% EtOAc /hexanes, and filtered through sintered glass. HPLC (Phenomenex Maxil 10 Si, 10 mm, 500 x 100 i. d., eluted in 4% (v/v) EtOAc /Hexanes, flow rate at 9 mi./min, UV detection at 254 nm) gave 19-epi-curacin A (4.2 mg, 21% yield) which showed the following: [a]25D +1220 (CHCI3, c = 0.635); UV X, (MeOH) 242 nm (e = 25000); IR u max: 2928, 2867, 2853, 1614, 1441, 1379, 1096, 1080, 999, 964, 912 cm-1; 1H NMR (300 MHz, 55 C6D6) 6 6.38 (dd, 1H,J= 15, 10.8 Hz, H-8), 6.02 (d, 1H, J. 10.8 Hz, H-9), 5.87 (m, 1H, H-15), 5.61 (dd, 1H,J= 10.5, 9.0 Hz, H-3), 5.58 (dt, 1H,J= 15, 10 Hz, H­ 7), 5.45 (dt, 1H,J= 10.5, 7.1 Hz, H-4), 5.12 (ddt, 1H, J = 9.0, 8.7, 1.1 Hz, H-2), 5.06 (m, 2H, H-16), 3.18 (s, 3H, -OCH3), 3.09 (m, 1H, H-13), 3.09 (dd, 1H, J =10.8, 8.2 Hz, H-lb), 2.75 (dd, 1H,J = 10.7, 8.7 Hz, H-1a), 2.26 (m, 2H, H-14), 2.23 (m, 2H, H-11), 2.11 (m, 2H, H-6), 2.11 (m, 2H, H-5), 1.70 (s, 3H, H-17), 1.65 (m, 2H, H-12), 1.55 (m, 1H, H-19), 1.49 (m, 1H, H-21) 1.34 (dt, 1H,J= 8.6, 4.4 Hz, H-20a), 0.85 (d, 3H,J= 5.7Hz, H-22), 0.48 (m, 1H, H-20b); 13C NMR (75 MHz, C6D6) 8 170.84 (C18), 136.07 (010), 134.99 (C15), 131.03 (C3), 130.65 (C7), 130.59 (C4), 127.54 (C8), 125, 55.97 (- OCH3), 39.25 (C1), 37.69 (C14), 35.44 (C11), 32.81 (C6), 31.82 (C12), 19 (C9), 116.45 (C16), 79.59 (C13), 73.85 (C2) 27.81 (C5), 23.21 (C19), 17.74 (C22), 17.50 (C21), 17.34 (C20), 16.25 (C17); LR EIMS m/z (rel. int. %) 373 (5), 342 (13), 332 (13), 274 (13), 181 (27), 180 (100), 166 (30), 98 (20), 91 (24), 85 (16), 79 (28), 67 (14), 55 (12); HRFABMS (positive) obs. [M + H]+ at m/z 374.2534 (1.7 mmu error for C23H36ONS). Preparation of TRO-curacin A (2 0). A sample of curacin A (18 mg, 48.3 1.1mol) was dissolved in 1.5 mL of acetic anhydride and 3 drops of D20. The reaction mixture was stirred and allowed to stand at rt overnight. The solvents were removed under vacuum. The residue was diluted with 25 % (v/v) EtOAc /hexanes, and filtered through sintered glass. The filtrate was chromatographed on a Phenomenex Maxil 10 gm silica column (50 x 10 cm) 56 developed at 9 mL/min with 25 % (v/v) EtOAc /hexanes to obtain TRO-curacin A (7.6 mg, 17.6 gmol, 42 % yield). IR uma. (film) 3270, 2929, 1678, 1650, 1547, 1536, 1435, 1371, 1096, 963, 913 cm-1; [a]22D +580 (c 0.1, CHCI3); UV Xmax (MeOH) 240 nm (log E = 4.49); 'H NMR (C6D6, 300 MHz) 8 6.42 (dd, 1H, J =15.0, 10.8, H-8), 6.02 (d, 1H, J=10.8, H-9), 5.87 (m, 1H, H-15), 5.62 (dt, 1H, J = 15.0, 10.0, H-7), 5.48 (dt, 1H, J=10.5, 7.1, H-4), 5.2 (dd, 1H, J=10.5, 9.0, H-3), 5.10 (m, 1H, H-2), 5.07 (m, 2H, H-16), 3.18 (s, 3H, -OMe), 3.09 (m, 1H, H-13), 3.04 (m, 2H, H-1), 2.35 (m, 2H, H-5), 2.26 (m, 2H, H-14), 2.23 (m, 2H, H-11), 2.11 (m, 2H, H-6), 1.8 (m, 1H, H-19), 1.7 (s, 3H, H-17), 1.65 (m, 2H, H-12), 1.58 (s, 3H, Me-CO), 1.12 (d, J=6.1, 3H, H-22), 1.11 (m, 1H, H-20b), 0.96 (m, 1H, H-21), 0.66 (m, 1H, H-20a); 13C NMR (C6D6, 75 MHz) 8 196.95 (C18), 167.99 (Me- CONH), 136.19 (C10), 135.37 (C15), 132.89 (C4), 131.58 (C7), 129.21 (C3), 129.21 (C3), 127.0 (C8), 125.64 (C9), 116.75 (C16), 79.95 (C13), 56.29 ( -OMe), 47.40 (C2), 38.05 (C14), 35.77 (C11), 33.59 (C1), 33.09 (C6), 32.14 (C12), 28.16 (C5), 22.97 (C19), 19.39 (C21), 16.60 (C17), 16.15 (C20), 11.95 (C22); GC EIMS (% rel. int.) obs. [M] + m/z 433 (1), 318 (4), 259 (4), 200 (5), 185 (5), 161 (10), 119 (20), 111 (11), 105 (18), 91 (19), 85 (21), 84 (34), 83 (100), 79 (20), 55 (22); HRFABMS (positive) obs. [M+H]+ at M/Z434.2728 (C25H40NO3S, deviation of 0.1 mmu). Preparation of the methyl thio ether derivative of curacin A (2 1). A solution of the TRO-curacin A (7.0 mg, 19 pmol) in dry THE (2 mL) was added into MeLi (1 gL, 34.1 gmol) at - 78 °C. In 1.5 h, Mel (1.3 gL, 20.4 limo') was 57 added into the mixture, and the solution was stirred at - 78 °C for 1 h. The solvents were removed under vacuum, and the residue was diluted with 100 % EtOAc and filtered through sintered glass. The filtrate was chromatographed on a Phenomenex Maxil 10 gm silica column (50 x 10 cm) developed at 9 mL/min with 100 % EtOAc. The first peak was repurified with 30 % EtOAc /hexanes to obtain the methyl thio ether derivative of curacin A (0.4 mg, 1.1 gmol, 5.7 % yield). 1H NMR (C6D6, 300 MHz) 8 6.40 (dd, J = 14.9, 10.7, 1H, H-8), 6.02 (d, 1H, J =10.7, H-9), 5.88 (m, 1H, H-15), 5.60 (m, 1H, H-7), 5.52 (m, 1H, H-4), 5.19 (dd, J=10.4, 9.0, 1H, H-3), 5.10 (m, 1H, H-16), 5.10 (m, H, H-2), 4.70 (br, 1H, NH), 3.18 (s, 3H, -OMe), 3.09 (m, 1H, H-13), 2.46 (d, J =5.8, 2H, H-1), 2.40 (m, 2H, H-5), 2.25 (m, 4H, H-14, 11), 2.18 (m, 2H, H-6), 1.91 (s, 3H, -SMe), 1.69 (s, 3H, H-17), 1.65 (m, 2H, H-12), 1.55 (s, 3H, Me -CO); 13C NMR (DEPT 135°, C6D6, 75 MHz) 8 135.37, 132.88, 131.43, 129.51, 127.67, 125.56, 116.81, 79.96, 56.32, 45.70, 39.89, 38.06, 35.80, 33.12, 32.16, 28.31, 22.94, 16.62, 15.89; GC EIMS (`)/0 rel. int.) obs. [M] + m/z 365 (1), 333 (12), 318 (2), 304 (1), 233, (8), 234 (9), 227 (14), 213 (25), 207 (18), 185 (38), 171 (22), 161 (73), 159 (43), 145 (40), 133 (46), 119 (100), 117 (48), 111 (55), 105 (92), 94 (32), 85 (89), 77 (42), 69 (66), 55 (47). Preparation of the curadisulfide A (2 2). The TRO curacin A (20) (14.0 mg, 38 gmol) in Me0H (2 mL) was treated with 0.5 M NaOH. After being stirred at rt overnight, the reaction mixture was acidified to pH 3 by adding 5N HCI. The solvents were removed under vacuum, and the residue was solubilized 58 with 100 `)/0 EtOAc and filtered through sintered glass. The filtrate was chromatographed on a Phenomenex Maxil 10 µm silica column (50 x 10 cm) operated at 9 mUmin with 90 % EtOAc /hexanes to obtain curadisulfide A (7.6 mg, 10.8 gmol, 54 % yield). 1H NMR (C6D6, 300 MHz) 8 6.42 (dd, 1H, J =15.0, 10.8, H-8), 6.02 (d, 1H, J=10.8, H-9), 5.87 (m, 1H, H-15), 5.62 (dt, 1H, J = 15.0, 10.0, H-7), 5.52 (m, 1HH-4), 5.48 (m, 1H, H-3), 5.25 (m, 1H, H-2), 5.12 (m, 2H, H-16), 3.21 (m, 1H, H-lb), 3.20 (s, 3H, -OCH3), 3.09 (m, 1H, H-13), 2.75 (dd, J = 13.5, 6.5, 1H, H-1a), 2.35 (m, 2H, H-5), 2.26 (m, 2H, H-14), 2.23 (m, 2H, H-11), 2.11 (m, 2H, H-6), 1.78 (s, 3H, Me-CO), 1.71 (s, 3H, H-17), 1.65 (m, 2H, H-12). 59 CHAPTER III MICROCOLIN C, A NEW ANTINEOPLASTIC LIPOPEPTIDE FROM THE MARINE CYANOBACTERIUM LYNGBYA MAJUSCULA ABSTRACT A new cytotoxic peptide, microcolin C along with three known compounds, microcolins A, B, and D, were isolated by bioassay-guided fractionation of the organic extract from Curacao and Granada collections of Lyngbya majuscula. The structure of microcolin C was assembled on the basis of extensive 2D-NMR spectroscopy, mass spectrometry, and comparison with the data sets for microcolins A and B. The absolute stereochemistry of microcolin C was deduced from spectral data analysis and chemical interconversion with microcolin B. Microcolin C was found to have an interesting profile of cytotoxicity to human cancer-derived cell lines. 60 INTRODUCTION Only few species rival the marine cyanobacterium Lyngbya majuscula in the structural diversity and biological activity of its secondary metabolites.76 For example, various collections of L. majuscula have yielded such unique metabolites as (+)-a- (S)-butyramido-y-butyrolactone (1), a C8 metabolite containing five-membered lactone,n malyngamide A (2), a chlorine-containing amide metabolite,18 and ypaoamide (3), a new herbivore antifeedent compound.79 Interestingly, most of these diverse structures may be considered as lipopeptides, pointing out a general trend in L. majuscula secondary metabolism. Recently, the advent of organ transplantation has resulted in new medical breakthroughs. This revolutionary event has been particularly due to the availibility of immunosuppressive agents such as cyclosporin A (CsA) (4),80 FK506 (5),81 and rapamycin (6).82 Although these compounds were isolated in the early 1970s, it was not until the 1980's that useful immunosuppressive properties were recognized. Cyclosporin A (4) is a cyclic undecapeptide which was isolated from the fungus Tolypocladium inflatum Gams. Cyclosporin A (4) is a lipophilic compound, and thus, cyclosporine must be solubilized for clinical administration. FK506 (5) is a 23-membered macrolide antibiotic which was extracted from a fermentation broth of the soil microorganism Streptomyces tsukubaensis. Rapamycin (6) is produced by Streptomyces hygroscopicus, and 61 structurally, it resembles that of FK506. The mechanism of action of cyclosporine (CsA) and FK506 is that both agents inhibit Ca2+ dependent signaling pathways in many cell types. Alternatively, rapamycin inhibits Ca2+ independent signaling pathways.83 -84 While cyclosporin A (CsA), FK506, and rapamycin are either used clinically or hold promise for their role in the prevention of organ transplant rejection and treatment of autoimmune disorders, there is still a critical need for new agents in this pharmacological class. Our recent interest in this regard has 0 OCH3 0 62 focused on the microcolin metabolites ,85-88 which are of particular note due to their extremely potent immunosuppressive activity. 0 4 63 RESULTS AND DISCUSSION In the course of investigating a Curacao collection of Lyngbya majuscula, the crude organic extract of this species was found by TLC to show several major metabolites with typical peptide characteristics (e. g., no H2SO4 charring reaction). This crude extract also was active in the brine shrimp toxicity assay. Using brine shrimp bioassay-guided fractionation of this extract (Figure 111.1), we isolated a new metabolite, microcolin C (9) (2.0 mg, 0.05 % of extract) as well as two known compounds, microcolins A (7) (140 mg, 3.2% of extract), B (8) (64.4 mg, 1.5% of extract). Also, continuing search for the microcolin type chemistry into a Granada collection of L. majuscula resulted in the isolation of microcolin D (10) (2.8 mg, 0.14%) (Figure 111.2). The existence of a hydroxyl group in 9 was suggested by an IR absorption at 3200-3700 (br) cm-1. The molecular formula was determined as C37H6307N5 by HRFABMS. The LRFABMS of microcolin C (9) showed fragmentation patterns similar to that of microcolin A (7). A careful analysis of these patterns revealed that the molecular ion as well as several fragment ion peaks of 9 lacked one (-Ac) group as compared to 7 (Figure 111.3). These data, together with the 1H NMR spectra of compound 9, suggested that it was a deacetyl derivative of microcolin B (8). The 1H NMR spectrum of microcolin C (9) (Table 111.1) was very similar to those of microcolins A (7)and B (8) (Figure 111.4). For example, the two singlets at 8 2.93 and 3.06 arising from two N-MeLeu and N-MeVal, the signals 64 Lyngbya (NAC 13 Aug/94-02), WG-EXT- 595, 4.39 g 52.8 mg saved 4.34 g VLC I I 4% E/H 4% E/H 595A 595B 25% E/H 595C I 50% E/H 50% E/H 595D 595E 85% E/H 595F (414 mg) 390 mg B S Toxic 30% E/M 100% M 100%E 595G 595H 5951 VLC 20 mg saved 60% E/H 595F-1 80% E/H 595F-2 (365 mg) 100% E 595F-3 Column Chromatography B S Toxic 5% M/CHCI3 10% M/CHCI3 50% M/CHCI3 595F-2A 595F-2B 595F-2C 595F-2D (299 mg) HPLC (C-18, 80% ACN/H20) B SIToxic 595F-2B1 595F-2B2 595F-2B3 595F-2B4 595F-2B5 595F-2B6 595F-2B7 (140 mg) (1.9 mg) (1.0 mg) Microcolin A (7) Microcolin C (9) B S Toxic (4 ng/ml) Figure III. 1 B S Toxic (200 ng/ml) (64.4 mg) MicrocolinB (8) B S Toxic (400 ng/ml) Flow chart for the isolation of microcolins (BS = brine shrimp) 65 Lyngbya (GMC-28 Jul/95-01) 1.96 g VLC [ I I I I 5% E/H 10% E/H 20% E/H 30% E/H 40% E/H 60% E/H 80%E/H 100% E G 100% M H L-y.----./ (103 mg) B S Toxic HPLC (C-18, 80% CH3CN/H20) I I (13 mg) G-1 (2.8 mg) G-2 1 (29 mg) (22 mg) (18 mg) G-3 G-4 G-5 Microcolin 0 (10) Microcolin A (7) MicrocolinB (8) Figure 111.1 Continued 66 H3 30 0 17 18 H3 31 15 22 CH3 CH3 CH3 0 44 34 43 6 H3 29 H3 42 H3 CH3 0 H N 19CH3 0 24 OAc H3 8 30 CH3 0 H3C' 40 34 18 28 17 H3 29 H3 CH3 0 H 27 15 33 N 26 N' 14 I CH3 CH3 CH3 0 I 42 41 32 19CH3 0 H3 6 H3 23 OH 24 9 Figure 111.2 Structures of microcolins A-D (7-10) 67 A 425 B 425 C 383 D 383 0 N N CH3 CH3 CH3 0...; A 282 H3 B 282 C 282 D 282 CH3 0 OR1 A 538 B 538 A 650 A 747 B 634 B 731 C 496 C 592 C 689 D 496 D 608 D 705 Microcolin A (7) R1 = Ac, R2 = OH Microcolin B (8) Microcolin C (9) R1 = Ac, R2 = H2 R1 = H, R2 = H2 Microcolin D (10) R1 = H, R2 = OH Figure 111.3. Mass fragmentations for microcolin isomers resonating at 5 4.5-5.5 attributed to the a protons of the amino acid residues, and a downfield doublet at 5 6.96 corresponding to an exchangeable NH proton, all resembled those of 7 and 8. The most significant feature differentiating 9 from 7 and 8 was the chemical shift (5 4.08) of the H-23 methine proton, and the loss of the acetate methyl group, indicating the loss of the acetate ester in 9. In addition, the a hydrogen (H-26) at 5 5.21 and 13 hydrogens (H-28) at 5 1.66 of the leu residue in 9 showed triplet and dd splitting patterns, while the corresponding hydrogens in 7 and 8 were a dd and ddd 68 pattern, respectively. Apparently, the absence of the acetyl moiety in microcolin C allows for rotational freedom of the isobutyl group of the leu residue. The analysis of 1H-1H COSY, HMBC, and TOCSY spectra of microcolin C (9) revealed the presence of the structural units, 4-methylpyrrolidinone, proline, N-Me-valine, threonine, N-Me-leucine, and dimethyloctanoic acid amide. The presence of 4-methylpyrrolidinone was determined by 1H-1H COSY correlations (cross peaks: H-2/H-3, H-2/H-4, H-3/H-4, H-4/H-6) and a comparison of chemical shifts with the proton and carbon spectra of microcolins A (7) and B (8). The proline, N-Me-valine, threonine, N-Me-leucine units were defined based on 1H-1H COSY correlations (proline: H-8/H-9, H-9/H-10, H-10/H-11, N- Me-valine: H-14/H-16, H-16/H-17, H-16/H-18, threonine: H-21/H-23, H-23/H-24, and N-Me-leucine: H-26/H-28, H-28/H-29, H-29/H-30, H-29/H-31) and TOCSY cross peaks (Figure 111.5) (proline: H-8 (8 5.47) - H-9 (8 2.40, 8 1.87) - H-10 (8 2.00) H-11 (8 3.80, 6 3.70), N-Me-valine: H-19 (8 3.09) 2.29) H-17 (8 0.81) - H-18 (8 1.01), threonine: H-22 (8 6.96) - H-21 (8 4.75) - H­ 23 (8 4.08), N-Me-leucine: H-32 (82.96) - H-26 (8 5.21) 1.43) - H-30 (80.89) H-14 (8 5.02) H-16 (8 H-28 (8 1.66)- H-29 (8 H-31 (8 0.94)). Although the cross peak corresponding to the Thr-Me (8 1.05) in the TOCSY was not observed, the Me resonance at 8 1.05 (H-24) showed strong COSY correlation to the Thr-13 proton resonance (8 4.08) indicating that this Me group belonged to the Thr component. The presence of the dimethyloctanoic acid amide moiety (C33 C42) was established by 1H-1H COSY and LRFABMS observing a fragment ion peak at 69 Figure 111.4 Comparison of 1F1 NMR spectra of microcolins A, B, and C (7-9) at 300 MHz in CDCI3. 7.0 6.5 7.0 6.5 3,5 4.5 6.0 5.5 5.0 4.5 4.0 01714 3. 65 60 5'5 4.5 2.5 1.0 2.5 1.0 1'11 70 No C.1 I 0 0. 0 C0 OC) 0 0 OI i 1 .1, 10­ (7 o' 6 o as 00 @ o 0 00 0 0 0 3 0 0 0 OCO 4 0 Val-a4 o 0 Leu dif.) -5 0-1-11 P 0 id -6 4 Thr-a I PPm 7 6 5 -7 (1 IT AI 3 , '" ppm I 2 I 1 Figure III. 5 TOCSY contour plot of microcolin C (9) at 600 MHz (CDCI3). 71 Table 111.1. 1H and 13C NMR data for 9 and 10 Microcolin C (9) Microcolin D (10) Position 1H (J in Hz)a 1 2 3 4 6 7 8 9 (a) (b) 10 11 (a) (b) 13 14 16 17 18 19 6.08 (dd, 6.0, 1.7) 7.23 (dd, 6.0, 1.7) 4.75 (m) 1.47 (d, 6.7) 5.47 (dd, 8.5, 4.8) 1.87 (m) 2.40 (m) 2.00 3.70 (m) 3.80 (m) 5.02 (d,11.0) 2.29 (m) 0.81 (d, 6.7) 1.01 (d, 6.6) 3.09 (s) 20 21 22 23 24 25 26 28 29 30 31 32 33 34 35 (a) (b) 36 37 (a) (b) 38 39 40 41 42 4.75 (m) 6.96 (d, 9.3) 4.08 (qd, 6.4, 1.9) 1.05 (d, 6.3) 5.21 (dd, 7.8, 7.8) 1.66 (dd, 7.6, 7.6) 1.43 (m) 0.89 (d, 6.7) 0.94 (d, 6.7) 2.96 (s) --­ 2.80 (m) 1.12 (m) 1.87 (m) 1.33 (m) 1.07 (m) 1.25 (m) 1.24 (m) 1.25 (m) 0.88 (t, 6.8) 1.10 (d, 6.7) 0.82 (d, 6.5) 13Cb 169.0 125.5 153.9 58.1 17.2 171.9 60.1 28.9 24.6 48.0 --­ 168.4 59.4 27.6 18.4 19.0 30.8 172.8 52.2 --­ 67.3 18.9 171.4 53.9 36.1 24.9 21.8 23.2 30.5 178.2 33.9 41.9 --­ 30.8 37.1 --­ 29.2 22.9 14.1 18.3 19.6 1H (J in Hz)c 6.04 (dd, 6.0, 1.8) 7.24 (dd, 6.0, 1.8) 4.76 (m) 1.41 (d, 6.7) 5.60 (dd, 8.6, 4.9) 2.00 (m) 2.45 (m) 4.35 (m) 3.92 (m) 3.76 (m) 4.98 (d,11.0) 2.25 (m) 0.77 (d, 6.7) 0.99 (d, 6.7) 3.05 (s) 4.70 (m) 6.99 (d, 9.2) 4.06 (qd, 6.4, 1.9) 0.95 (d, 6.3) 5.16 (dd, 7.8, 7.8) 1.62 (dd, 7.6, 7.6) 1.39 (m) 0.82 (d, 6.7) 0.89 (d, 6.7) 2.94 (s) --­ 2.80 (m) 1.12 (m) 1.83 (m) 1.26 (m) 1.07 (m) 1.24 (m) 1.22 (m) 1.23 (m) 0.83 (t, 6.9) 1.08 (d, 6.8) 0.82 (d, 6.5) 13Cd 169.9 125.3 154.2 58.1 16.9 174.6 58.5 36.5 - 71.9 57.1 -­ 169.2 59.3 27.4 18.3 18.8 30.6 172.8 52.1 - -­ 67.2 18.9 171.3 53.8 36.0 24.9 21.8 23.1 30.5 178.2 125.5 41.8 30.7 37.0 29.1 22.9 14.1 18.2 19.5 a300 MHz in CDCI3, b75 MHz in CDCI3, C600 MHz in CDCI3, d150 MHz in CDCI3 72 m/z 282 which is consistent with the same linear portion as 7 and 8. Complete assignments of these partial structures were accomplished by HMBC. The methine proton resonance at S 4.75 (H-4) showed a HMBC correlation to C-7 (8 171.9) indicating that the pyrrolidinone and proline groups formed an amide linkage. HMBC cross peaks between a methine signal at 8 5.47 (H-8) and C-13 (8 168.4), between methylene signals at 8 3.80, and 8 3.70 (H-11) and C-13 (8 168.4) established a connection between a proline and valine unit through an amide bond. Similarly, HMBC correlations between a methine proton at 8 5.02 (H-14) and C-20 (8 172.8), between a methine resonance at 8 4.75 (H-21) and C-25 (8 171.4), between a methine peak at 8 5.21 (H-26) and C-33 (8 178.2) determined the linkages of the valine-threonine-leucine-dimethyloctanoic acid amide residues. The absolute stereochemistry of 9 was deduced to be 4S, 8S, 14R, 21S, 26R, 34R, and 38R, identical to that of 8 whose absolute configuration was assigned in a recent study.89 In fact, microcolin C could be synthetically produced through the enzymatic hydrolysis90-92 of microcolin B (Figure 111.6). 3 H3 CH 0 H3C 0 Esterase /\/\/\AN N E HZ\ ./\(\)\ 3cr,2 Phosphate buffer CH3 orb cm, ocA cc m3 o E E N I CH3 CH3 CH3 0 A CH3 0 0 8 'OH H30- 9 Figure 111.6 Enzymatic hydrolysis of microcolin B 73 Table 111.2. Tumor cell growth inhibitory activity and cytotoxicity of microcolin C in the National Cancer Institute in vitro 60-cell line screen. Panel/Cell line Log 10GI50a Logi oTG 1 b Log10LC50c Leukemia CCRF - CEM MOLT-4 SR < -8.60 < -8.60 < -8.60 < -8.60 > -4.60 -8.09 < -8.60 > -4.60 Non-Small Cell Lung Cancer NCI-H322M < -8.60 < -8.60 -8.51 < -8.60 < -8.60 -8.47 < -8.60 < -8.60 < -8.60 -8.08 -8.08 -7.94 -7.76 -8.18 -5.50 < -8.60 -8.59 -8.05 < -8.60 -7.96 -5.13 < -8.60 -8.59 -8.40 -7.66 -6.13 -5.76 -7.85 0.75 2.75 -6.65 1.95 4.00 -5.55 2.96 Colon Cancer HCT-15 HT29 KM12 SW-620 Melanoma LOX IMV1 Prostate Cancer PC-3 Breast Cancer MDA-MB-231/ATCC HS 578T MG MIDd Deltae Range( -4.98 3.91 aGI50, concentration required for 50% growth inhibition; bTGI, concentration required for total growth inhibition; cLC50, concentration required for 50% lethality; dMG-MID, the calculated mean panel GI50 TGI, or LC50; eDelta, the number of log10 units by which the value of the most sensitive line(s) of the panel differs from the corresponding MG-MID; (Range, the number of log10 units by which the value of the most sensitive line(s) of the panel differs from the value of the least sensitive lines.93 i 4 3 2 Figure 111.7 11-1 spectrum of microcolin D (1 0) at 600 MHz (CDC13). 1 TTTTTTT ppm 6 5i 4 3 Figure III. 8 1H-1H COSY contour plot of microcolin D (1 0) at 600 MHz (CDCI3). 76 Treatment of compound 8 with porcine liver esterase (PLE) in phosphate buffer gave compound 9 which was confirmed to be identical to the natural product microcolin C by comparison of optical rotation, mass spectra, and 'H and 13C NMR spectra. Desacetylmicrocolin A (10) was previously prepared as a semi-synthetic derivative by the Harbor Branch Oceanographic Research Group' as part of their evaluation of the immunosuppressive activity of this series. Comparison of NMR (Figure 111.7 and 8) and mass spectral data for compound 10 isolated from L. majuscula and the semi-synthetic desacetyl microcolin A showed them to be identical. Microcolin C (9) was investigated in the National Cancer Institute in vitro 60-cell line screen. Several cancer cell panels showed growth inhibition (Table 111.2). Microcolins A-C (7-9) also exhibited potent toxicity to brine shrimp (LC5O of 0.004 gg/ml, 0.4 pg /ml, and 0.2 µg /ml, respectively). EXPERIMENTAL General Experimental Procedures. NMR spectra were recorded on Bruker DRX 600, AM 400, and ACP 300 spectrometers in the solvent specified. Chemical shifts were referenced to solvent CDCI3 signal at 7.27 ppm for 1H and 77.0 ppm for 13C NMR. Mass spectra were recorded on Kratos MS 50 TC and Finnigan 4023 mass spectrometers. Gas chromatography/mass spectrometry 77 (GC/MS) was carried out utilizing a Hewlett-Packard 5890 Series II GC connected to a Hewlett-Packard 5971 mass spectrometer. UV spectra were obtained on a Hewlett-Packard 8452 A UV-VIS spectrophotometer and IR spectra on a Nicolet 510 spectrophotometer. All solvents were distilled from glass prior to use. Biological Material. The cyanobacterium L. majuscula was collected from shallow water (0.5 - 2.1 m) on August 13, 1994, at CARMABI, Curacao, Netherlands Antilles, and stored in i-PrOH at low temperature until work-up. A voucher specimen is available from W.H.G. (collection NAC-13 Aug 94-2). Extraction and Isolation. A total of 305 gm (dry wt) of Lyngbya majuscula was extracted with CHCI3/MeOH (2:1 v/v) three times to give 4.39 gm of crude extract. Gradient vacuum chromatography of the crude extract with EtOAc /hexanes gave a brine shrimp toxic fraction containing the microcolins (414 mg). This fraction was subjected to vacuum liquid chromatography on silica (60%-100% EtOAc /hexanes) to give brine shrimp toxic active fraction (365 mg) which was further chromatographed on silica (5%-50% Me0H/CHC13) to yield 299 mg. Finally, this brine shrimp toxic fraction (5% Me0H/CHC13) was fractionated by preparative C18 HPLC (Hibar LiChrosorb RP-18,7 jim 250 x 10 mm) and eluted with 80% CH3CN/H20 using UV detection at 254 nm to yield microcolins A (140 mg, 3.2% of extract), B (64.4mg, 1.5% of extract), and C (2.0 mg, 0.05% of extract). 78 Microcolin C (9): Microcolin C was isolated as a colorless oil and showed the following: [a]25D -169° (c 0.5, EtOH); UV km (MeOH) 210 nm (E = 32000); IR um (Film): 3200-3700 (br), 2935, 2335, 1735, 1645, 1250 cm-1; LR FABMS obs. M+ at m/z (rel. int. %) 689 (1.1), 592 (1.0), 496 (22.7), 478 (15.2), 383 (4.5), 365 (4.5), 282 (67), 100 (100); HR FAB MS (positive ion) obs. [M + H]+ at m/z 690.4807 (0.2 mmu error for C37H6407N5). Enzymatic Hydrolysis of Microcolin B (8). Compound 8 (6 mg, 8.2 p.mol) was dissolved in methanol (0.1 mL) and the resulting solution was added to 0.2 M phosphate buffer (6 mL) containing porcine liver esterase (PLE, 0.5 mg, 100 units Sigma). The solution was stirred for 5.5 hr at room temperature and then repeatedly extracted with ether (3 x 10 mL). The extracts were dried over MgSO4, the solvent removed in vacuo, and the crude product subjected to preparative C18 HPLC (Hibar LiChrosorb RP-18, 7 p.m 250 x 10 mm, 80% CH3CN/H20, UV detection at 254 nm) to yield microcolin C (4.3 mg, 72% yield); [a]25D -175° (c 1.1, EtOH); UV 'max (MeOH) 210 nm (E = 33000). 79 CHAPTER IV VIDALENOLONE, A NOVEL SECONDARY METABOLITE FROM THE INDONESIAN MARINE RED ALGA VIDALIA SP. ABSTRACT A new secondary metabolite, vidalenolone, was isolated from an Indonesian red alga Vidalia sp. The structure of this new cyclopentenolone­ containing compound was determined by a combination of spectroscopic methods. 80 INTRODUCTION Approximately 5500 species of red algae (Rhodophyta) have been taxonomically identified.' Many red algae have been recognized as prolific producers of halogenated monoterpenes and brominated phenols.95 Halomon (1), a novel antitumor metabolite, and related compounds (2-8),96 isolated from the red alga Portieria homemannii from different geographical locations, are good examples of bioactive terpenoids. Although these halogenated monoterpenes look structually simple, the structure determination of these compounds is not due to the difficulty in obtaining good mass data and the difficulties in placing halogen substituents. CI CI 2 ci 3 CI CI 6 CI 7 CI CI 81 In contrast to other genera of red algae, there has been relatively little exploration of secondary metabolites from the genus Vidalia. Only a few compounds have been found in this genus. These include the antiinflammatory vidalols A (9) and B (10),97 a new amino acid (11),98 2-amino-5 trimethylammoniopentanoate, and a halogenated cyclopentenediol (12)." In our continuing search for bioactive compounds from Indonesian marine red algae, we have isolated a new metabolite, vidalenolone (13) (2.0 mg), from Vidalia sp. Here, we report the isolation, structure determination and biogenesis of vidalenolone (13). OH HO HO HO OH OH HO OH OH 10 9 NH2 /CH3 + ....,..........,..--......-N-CH3 00C 0H3 11 82 RESULTS AND DISCUSSION A collection of Vidalia was extracted with CHCI3 and Me0H (2:1 v/v) and the resulting crude extract was fractionated by Si gel vacuum liquid chromatography. The fraction eluting with 80% EtOAc /hexanes was further chromatographed to give fractions which were finally purified by normal phase HPLC to yield a new cyclopentenolone compound 13 . The molecular formula was determined to be C13H1404 by HR CIMS (obs. [M + H]+ at m/z 235.0971, A 0.1 mmu). Hence compound 13 possessed seven degrees of unsaturation, four of which were due to a phenolic ring, one due to an olefin, and one due to a ketone functionality. The remaining one degree of unsaturation required 13 to contain one ring. The IR spectrum showed the existence of a hydroxyl group (3450 cm-1), and an a, 13 - unsaturated carbonyl group (1625 cm-1). The presence of a para­ hydroxybenzyl moiety was deduced from the positive CI mass spectrum which gave a major peak at m/z 107, compatible with C7H70. This was supported by 1H NMR data 8 6.9 (2H, d, J = 8.3 Hz), 6.6 (2H, d, J = 8.3 Hz) (Figure IV.1) and 13C NMR data (8 155, 131 (2C), 125, 114 (2C)). The 1H NMR spectrum in DMSO-d6 revealed signals for two methylenes (8 2.42, 2.71), one 0-methyl group (8 3.05), a methine (8 6.3), four protons in the aromatic region, and two deshielded hydroxyl signals (8 9.2, 9.5) (Table IV.1). Additionally, the 13C NMR was in good agreement with the presence of a carbonyl carbon at C-1 i Ppm u I 9 I 8 I 7 6 I 1 5 1 4 1 3 2 Figure IV.1 11-I NMR spectrum of vidalenolone (13). 1 84 Table IV.1 11-I and 13C NMR data for vidalenolone (13) Position 1H (J in Hz)b 13cc 1 202.79 2 153.00 HMBC correlations 3 6.30 (t, 3.1) 129.66 Cl, C4, C5 4 2.40 (dd, 5.7, 3.0) 30.19 Cl, C3, C5 5 80.37 6 2.69 (d, 5.6) 7 40.72 C1, C3, C4, C5, C7, C8 125.63 8 6.9 (d, 8.3) 131.09 C6, C7, C9, C10 9 6.6 (d, 8.3) 114.70 C7, C8, C10 10 155.00 11 ( -OCH3) 3.05 (s) 50.95 C4, C5 (H-O-Ph-) 9.25 C9, C10 (H-0-) 9.55 C1, C2, C3 a Spectra were recorded in DMSO-d6. b 1 1-1 NMR at 300 MHz, cl3C NMR at 75 MHz 85 OH 0 n c b a Figure IV.2 Partial structures of vidalenolone (13) OH OH Figure IV.3 Structure of vidalenolone (13) d 86 (8 202.79), a polarized olefin (8 153.13, 129.66), a methoxy carbon (8 50.95), and two methylene carbons (8 40.72, 30.19). Detailed spectral analysis of 13 provided the partial structures, a, b, c and d (Figure IV.2). The partial structure a was deduced from 1H-1H COSY correlations between a methine proton at 8 6.9 (H-8) and 8 6.6 (H-9). The methylene proton resonance at 8 2.69 (H-6) showed HMBC correlations to C-7 and C-8 suggesting its connection to a hydroxylbenzyl moiety. The methylene proton resonance at 8 2.40 (H-4) in the structural fragment b showed COSY correlation to a methine resonance 8 6.30 (H-3). The deshielded 13C chemical shift of the quaternary carbon resonance 8 153.00 (C-2) indicated that it bore the free hydroxyl group. The location of this free hydroxyl group was further supported by HMBC correlations to C-1, C-2, and C-3. The partial structure c was defined based on the methoxy proton resonance 8 3.05 (H-11) showing HMBC correlation to a deshielded quatenary carbon resonance 8 80.3 (C-5). The carbon chemical shift at 8 202.79 (C-1) indicated the presence of the ketone functionality (partial structure d). Complete assignments from these partial structures were achieved by further HMBC analysis. The methylene protons at 8 2.69 (H-6) showed HMBC correlations to C-5 connecting partial fragments a and c. HMBC correlations between methylene protons at 8 2.40 (H-4) and C-5, between a methine proton at 8 6.30 (H-3) and C-5 enabled 87 connection of b and c. Finally, HMBC corelations between methylene protons 8 2.69 (H-6) and C-1, and a hydroxyl proton 8 9.55 and C-1 completed the planar structure of vidalenolone (13) (Figure IV.3). The stereochemistry of 13 is yet to be established. Stereochemical study of 13 was attempted using CD analysis. However, the results were not conclusive because of an abnormality of observed peaks. The biogenetic origin of vidalenolone (13) is presented in Figure IV.4. Tyrosine (14) and succinyl - CoA (17) could be reasonable major precursors in this biosynthetic pathway. Similar condensation of succinyl-CoA and hydroxyketone was proposed for the assembly of a multifunctional mC7N moiety of the munumycin group of antibiotics.lw In this proposed pathway, tyrosine may be transformed by the action of transaminase to a-keto acid (15). Subsequently, the carboxylic acid in 15 could be reduced to the corresponding alcohol (16) which further reacts with succinyl-CoA (17) to form a new carbon-carbon bond (18). Five- membered ring formation can take place by decarboxylation of (18) to give the a, 13-dihydroxyketone (19). The tent- hydroxyl group of 19 could undergo 0­ methylation to yield 20. Subsequently, the a-hydroxyl of 20 could be oxidized to give the diketone (21) which enolizes to form the final product, vidalenolone (13). 88 OH OH HO 15 14 SCoA H3C HO v OH 0 20 HO HO 19 OH HO HO 21 13 Figure IV.4 Proposed biogenetic pathway of vidalenolone (13) 89 EXPERIMENTAL General Experimental Procedures. NMR spectra were recorded on a Bruker ACP 300 spectrometer in the solvent specified. Chemical shifts were referenced to the solvent DMSO signal at 2.50 ppm for 1H and 39.51 ppm for 13C NMR. Mass spectra were recorded on Kratos MS 50 TC and Finnigan 4023 mass spectrometers. UV spectra were obtained on a Hewlett-Packard 8452 A UV-VIS spectrophotometer and IR spectra on a Nicolet 510 spectrophotometer. CD measurement was obtained on a Jasco 41A spectropolarimeter. All solvents were distilled from glass prior to use. Plant Material. The marine red alga Vidalia sp. (Rhodophyta; Rhodomelaceae; Ceramales) was collected on November 5, 1994, at Ang Island, Indonesia. It was found growing subtidally on volcanic substrate. The alga was preserved in i-PrOH at low temperature until extraction. A voucher specimen (collection IAI-5-Nov-94-1) is stored in the natural products laboratory of the College of Pharmacy, Oregon State University. Extraction and Isolation. The i-PrOH preserved alga Vidalia was extracted with CHCI3 and Me0H (2:1 v/v) three times at room temperature. The solvent was evaporated to dryness under reduced pressure at 30 °C to provide a crude extract (6.02 g) which was fractionationed by Si gel vacuum liquid column chromatography with a gradient solvent system (hexanes- EtOAc). The fraction eluting with 80% EtOAc /hexanes was further chromatographed to give 90 eight fractions. Fraction 5 was subjected to repetitive HPLC on a Al !tech Versapack Si column (10 gm, 2 x 300 x 4.1 mm) with 25% hexanes/ EtOAc to afford pure vidalenolone (13). Vidalenolone (13): [a]25D -950 (c 0.31, MeOH); UV Xmax (MeOH) 209 nm (E = 32000), 224 (E = 35000), 266 (E = 12000); IR uma, = (film) 3450 (br), 2935, 2335, 1700, 1625 cm-1, LR CIMS (rel. int. %) obs. [M + H]+ at miz 235 (18), 234 (13), 219 (2), 217 (2), 205 (18), 204 (21), 203 (100), 157 (6), 135 (6), 129 (45), 128 (90), 107 (78); HR CI MS (positive ion) obs. [M + H]+ at nilz 235.0971 (0.1 mmu error for C13F11404); CD (MeOH): AE = -3.3, -1.3, -1.8 (Am.), = 220, 250, 270 nm). 91 CHAPTER V. Analysis of Oxylipin Products from Gametophyte Cell Cultures of the Brown Alga Laminaria saccharina ABSTRACT Filamentous cells isolated from female gametophytes of the brown alga Laminaria saccharina were cultured in flask or bioreactor culture. These cultures produced a variety of 15-lipoxygenase metabolites. Analysis by GC­ MS of TMS ethers of the methylated hydroxy acids identified the presence of three oxylipins: 13-hydroxy-9,11-octadecadienoic acid (13-HODE), 13-hydroxy­ 6,9,11,15-octadecatetraenoic acid (13-HODTA), and 15-hydroxy-5,8,11,13­ eicosatetraenoic acid (15-HETE). Feeding experiments were carried out to attempt the enhancement of these 15-lipoxygenase metabolites using linoleic acid, a-linolenic acid, y-linolenic acid, arachidonic acid, jasmonic acid and calcium ionophore antibiotic. The presence of linoleic acid, y-linolenic acid and jasmonic acid increased the production of the three oxylipins 2 to 4 times over the controls. On the other hand, addition of antibiotic did not increase the yield, while a-linolenic acid suppressed production of these metabolites. Arachidonic acid addition gave rise to a variety of HETE products via autoxidation. 92 INTRODUCTION Macrophytic marine algae or seaweeds have proved to be a rich source of pharmacologically active compounds.' They are able to biosynthesize eicosanoids and related oxylipins through different pathways of the arachidonic acid (AA) cascade (see Figure 1.1 in Chapter 1).' In mammalian systems, precursors such as arachidonic acid lead to a number of pharmacologically important metabolites known cumulatively as eicosanoids which include the HETEs, prostaglandins, thromboxanes, hepoxilins, and leukotrienes.1' Many of the secondary metabolites isolated from marine macroalgae show potential as biomedicines. Of particular interest are marine oxylipins. Many of the bioactive oxylipins isolated from different classes of marine macroalgae are not found in mammals. This is especially true of some genera of red and brown algae. Marine oxylipins appear to be the product of the lipoxygenase pathway. For example, the red algae tend to metabolize C20 acids via the 12­ lipoxygenase pathway, green algae utilize C18 acids at C-9 and C-13, and brown algae metabolize both C18 and C20 acids as substrates. Figure V.1 represents a variety of oxylipins produced by various lipoxygenase and significant fragment ions from their derivatives as observed by GC-EIMS. A major problem in drug discovery is that the yields of secondary metabolites from natural sources are usually in such low amounts that an adequate supply can be serious a problem, not only for preliminary in vivo drug 93 02CH3 5-HETE [M]+ 406 8-HETE 1.- 225 11-HETE 9-HETE CO2CH3 CO2CH3 ..: , OTMS i 295 --ff' 12-HETE 14-HETE 15-HETE Figure V.1 Expected fragment patterns of various oxylipins cr ,ce \+ eb 01/ og- r';')4) '', / S 1/ 95 GO 2.0-,3 GO P-4,3 0020-A3 "0110:, 3eA.---- 1 3-IA001N \M1+ g78 311-*-­ A3-1-3tODE. ot NOTE klK 382 ov 380 CO2.04 96 evaluations, but also for the commercial market. There are several approaches which enable an adequate supply of bioactive compounds for commercial products. 102,103 The traditional approach is to recollect larger quantities of the organism. However, this approach has been frequently criticized by ecologists and environmentalists because ocean resources are often over utilized in the name of science.' For example, over 1 ton of the sea hare Dolabella auricularia was collected to obtain 29 mg of dolastatin 10, a potent anticancer compound.' Subsequently, to provide material for clinical trials, about 700 tons of the sea hare would be required. The anti-ovarian-cancer drug taxol offers another good example of the supply issue. The amount of tree bark from the Pacific yew tree needed to obtain 1 kg of pure taxol is about 30,000 pounds, while the annual medicinal need for treating patients in the U.S. is over 25 kg. 102 Another approach is to try the total synthesis of the desired target compound. One drawback of this approach is that it may not be suitable for some compounds due to the difficulty of controlling stereochemistry and the poor yield of the desired compounds. For example, a group at OSU reported the total synthesis of curacin A, however, with only a 0.6 % overall yield." A third approach is to culture and harvest the organism. This approach has been successfully carried out with culturable marine microorganisms in the laboratory. 107,108 But, the scale-up culture of a marine microorganism for clinical trials or pharmaceutical demands has not been reported." A relatively new approach is to utilize biotechnology such as bioreactors to cultivate marine organisms. Undifferentiated cells are placed in a liquid 97 suspension culture, followed by stimulation to yield the target secondary metabolites in the culture. 110,111 Advantages of this approach over traditional culture include: fast and controlled cultivation of cells in a bioreactor and the opportunity to increase the production of the desired compound through a fine level of control in changing the culture conditions."' However, there are few reports regarding cell culture studies using bioreactors. In this work, the brown alga L. saccharina was cultivated in a bioreactor and isolation, identification, and quantitative analysis of oxylipins was described. This work was a collaborative effort with the laboratory of Dr. Rorrer in the Department of Chemical Engineering at O.S.U. Dr. Rorrer's laboratory was responsible for the bioreactor design, and start-up and cultivation of L. saccharina female gametophyte cells."' RESULTS AND DISCUSSION This work describes the capability of the brown alga L. saccharina to produce bioactive marine oxylipins utilizing gametophyte cell suspension cultures. The procedures for the isolation and analysis of oxylipins produced from the L. saccharina female gametophyte cell biomass are shown in Figure V.2 and described in the experimental section. Prior to the quantitative analysis of the oxilipins produced in these cultures, the qualitative analysis was carried out using GC-MS. Fresh wet tissue was homogenized in 20 mL of CH2C12/Me0H (2:1 v/v) (Figure V.2). 98 Fresh wet tissue homogenize add internal standard pool extracts 1 Methylation of extract (CH2N2) Partial puification of oxylipins by prep-TLC scrape the spot containing desired compounds and internal standard 1 Trimethylsilyl (TMS) derivatization Hexamethyldisilazane (HMDS) Chlorotrimethylsilane (TMS-CI) Pyridine 1 vacuum 1 centrifugation GC-Mass spectrometry Figure V.2 Overview of the identification and analysis of oxylipins in L. saccharina cell culture 99 Ricinelaidic acid was added to the extract as an internal standard for quantitative analysis. The extracts were pooled, filtered, and resulting solvent was dried using a Buchi rotary evaporator. A portion was then methylated with CH2N2 in Me0H. This methylated extract was subjected to Prep-TLC using silica gel 60 F254 in 25% EtOAc /hexanes. The spot containing the desired methylated hydroxy acids was broadly scraped off the TLC plate. Several methylated oxylipins were used for references to locate the methylated hydroxy acid fraction of the extract. The resulting powdered gel was extracted with Et20 and evaporated by rotavaporation. Tissue extraction was derivatized with appropriate reagents to be TMS ethers of the methyl esters of the hydroxy fatty acids prior to the injection into GC-MS. Then, derivatized materials were analyzed by GC-MS (Figure V.3). TIC: 3- 107 -4.D 3 2000000­ 4 5 1500000­ 12 2 1000000 6 20 500000­ 11 13 16 mor10 ime-->7.00 8.00 9.00 10.00 1-; 11.00 12.00 13 00 14.00 Figure V.3. Total Ion Chromatogram (TIC) of TMS-hydroxy fatty acids produced from L. saccharina gametophyte cell suspension. 100 The significant peaks were identified by matching the averaged mass spectra for the eluted peak to mass spectra of standards using the library program of the HP Data station (Table V.1). Detailed analysis of GC peaks, especially, in the range of 12.6 - 15.7 min (Figure V.3), resulted in the identification of three oxylipins derived from action of a 15-lipoxygenase: 13-hydroxy-9,11-octadecadienoic acid (13-HODE) (1), 13-hydroxy-6,9,11,15-octadecatetraenoic acid (13-HODTA) (2), and 15­ hydroxy-5,8,11,13-eicosatetraenoic acid (15-HETE) (3). The identities of these TMS ether derivatives of the methyl esters of 13­ HODE (1), 13-HODTA (2), 15-HETE (3) were indicated by their distinct fragmentation patterns at m/z 311, 309, and 335 (Figure V.4 and 5), respectively, as well as matching their GC retention times to that of the corresponding authentic standard compounds. The yield of oxylipins in the sample was calculated by employing the internal standard method using the total peak areas of the corresponding oxylipin, the peak area of the standard (ricinelaidic acid), the concentration of the internal standard added to the analyzing sample, and the detector response factor. During the course of quantitative analysis of these oxylipins, two factors made this study challenging. First, the derivatized oxylipins rearranged during GC and showed separate peaks. This phenomenon of rearrangement has been commonly observed when these kinds of oxylipins are injected into a Gc.12 Second, the internal standard, ricinelaidic acid, co-eluted with 13­ 101 # Peak Quality (%)3 1 78 Palmitic acid Me ester 2 99 Palmitoleic acid Me ester 3 95 Palmitic acid Me ester 4 81 6,9,12-Octadecatrienoic acid, methyl ester 5 99 Oleic acid Me ester 6 99 Oleic acid Me ester 7 91 Si lane, [(3,7,11,15-tetramethy1-2-hexadecenyl)oxy]trimet 8 99 Docosane 9 93 Arachidonic acid Me ester 10 95 Methyl 15-HODTA, TMS ether 11 12 Si lane, dimethy1-2-propenyl(tetradecyloxy)­ 12 90 Ricinelaidic acid, TMS ether (Internal standard) + Me 13-HOD 13 53 Methyl 14-HEPE, TMS ether 14 91 Methyl 15-HODTA, TMS ether (rearranged) 15 73 Methyl 13-HODE, TMS ether 16 50 Hexanedioic acid, mono (2-ethylhexyl) ester 17 32 Pregn-17(20)-en-16-one, (5 alpha., 17Z)­ 18 95 Methyl 15-HETE, TMS 19 25 1,3,5-Triazine-2,4-diamine, 6-(3-methylpheny1)­ 20 64 Bis (2-ethylhexyl) phthalate 21 80 Methyl 15-HETE, TMS ether (rearranged) ID Table V.1. Identified compounds for the TIC chromatogram shown in Figure V.3 a Quality ( %) means purity of all m/z fragments of being analyzed compound relative to all m/z fragments of standard compound. 102 HODE (1) (Figure V.5). Thus, the detector response factors used to quantify 13­ HODE production were based on peak areas calculated from mass-selective or mass extract ions at m/z 311 for 13-HODE and at m/z 187 for the ricinelaidic acid. Detector response factors used to quantify 13-HODTA (2) and 15-HETE (3) were determined based on peak areas obtained from TIC data. In an attempt to enhance the yield of oxylipins derived from the 15­ lipoxigenase pathway, six precursor or elicitor feeding experiments were carried out. These included linoleic acid (6,9-octadecadienoic acid), a-linolenic acid (9,12,15-octadecatrienoic acid), y-linolenic acid (6,9,12-octadecatrienoic acid), jasmonic acid, antibiotic (Ca-ionophore), and arachidonic acid. The relationship of these precursors as well as several other and their corresponding 15-lipoxigenase products is depicted in Figure V.6. At the 20th day of cultivation, 2 mL of a 10 g/L stock solution containing a given precursor or elicitor was added to the liquid suspension culture. Then, the culture was incubated for an additional 10 days to give a total cultivation time of 30 days. The toxic effect of exogenously added precursors to cell suspension cultures was also examined. It was found that both the structure of the fatty acid and its concentration in the culture medium are critical to its toxicity. The general trend of toxicity for these fatty acids is a-linolenic acid > linoleic acid > y­ linolenic acid. The maximum tolerable precursor concentration for the culture was found to be 200 mg/L and concentrations over 1000 mg/L were toxic to 103 bundance Average of 12.747 to 12.769 min.: 3-105-7.D (+, 73 ) 309 CO2CH3 90001 8000 \OTIA4 309, 7000 13-HODTA [Mr 378 6000 5000 4000­ 3000 91 1291-45 2000­ 187 59 1000­ 49 0 /Z 243 219 , -> 50 100 150 Abundance 200 -AL 1 280 , 250 't ti 34'7363 I "' , , 300 350 , 400 #40: Methyl 15 -HETE, TMS (*) 7 3 90001 CO2CH3 80001 7000: 4bTMi'f 335 225 6000 15-HETE 5000­ 4000: 3000= 91 129 2000 155173 1000, 55 199 251 0 M/Z -> 50 100 150 200 250 335 316 291 349 300 350 391406 400 Figure V.4 Mass spectra and structures of TMS ether derivatives of the 13­ HODTA and 15-HETE 104 (A) Abundance #13: Methyl 13 -RODE, TMS Ether (*) 7 3 90001 CO2CH3 80001 70001 311. 130 6000: tTMiA 13 -NODE R,A1+382 50001 4000: 311 30001 225 93 20001 55 382 155171 10001 199 M/Z -> 111TH 50 100 (B) bundance 150 269 292 200 250 335 300 367 350 Average of 12.940 to 12.952 min.: 3-104-7.D 187 (+, 418 .,1., 400 ) 9000 1 CO2CH3 8000 1 OTMS 7000 1 187 6000 Ricineladcadd 5000 1 40001 73 3000 20001 103 270 311 1000: 55 129 0 4 50 1551711 44140'rmli-14"4 100 150 200 299 225239 . ,-" . 250 300 337 3682 r, 350 Figure V.5 Mass spectra and structures of TMS ether derivatives of (A) methyl 13-HODE (1) and (B) of co-eluted methyl ricinelaidic acid standard (4) and methyl 13-HODE. 105 cultures in the 10 day incubation. The production of 13-HODTA, 13 -NODE, and 15-HETE following addition of the six precursors or elicitors is shown in Table V.2 and Figure V.7 through Figure V.10. Based on absolute yields, all culture experiments produced 13 -NODE in the greatest quantity, followed in order by 13-HODTA and 15-HETE. Figure V.7 shows that the yield of 13-HODTA was significantly enhanced by the addition of y-linolenic acid, resulting in a yield over three times higher than that of controls. The yield of 13 -NODE was increased in three precursor feeding experiments (linoleic acid, a-linolenic acid, and y-linolenic acid). But, the enhancements were not highly significant as less than two times the value of the controls were observed (Figure V.8). The yield of 15-HETE was significantly increased by several of the feeding precursor experiments (Figure V.9). Cultures provided with arachidonic acid led to a significant increase in 15-HETE production. However, careful analysis utilizing extract ion chromatography in GC-MS revealed that the exogenous addition of arachidonic acid resulted in the production of various HETE's due to non-enzymatic autoxidation. This fact was established by observing significant ion fragments at m/z 295 (12-HETE), 265 (8-HETE), 255 (9-HETE)225 (11-HETE), and 335 (15-HETE). It is notable that the feeding experiments with a- linolenic acid gave yields of oxylipins (15-HODTA (2) and 13-HODE (1)) lower than control experiments. One possible explanation for this observation is that, from the toxic screening 106 Fatty Acid Precursor Product COOH COOH 15-LOX peroxidase ''OH linoleic acid (LA) COOH 13 -NODE (1) COON 15-LOX peroxidase OH H a-linolenic acid (a-LA) 13-HOTE COOH 15-LOX COOH peroxidase OH rlinolenic acid ( y-LA) 13-HOTE ( y) COON 15-LOX COOH peroxidase OH stearidonic acid 13-HODTA (2) OOH 15-LOX peroxidase arachidonic acid SOH 15-HETE (3) Figure V.6 Hydroxy fatty acid production from precursors by 15-LOX/peroxidase metabolism 107 13-HODTA 13-HODE 15-HETE #1. L s control 123.2 ± 39.4 43.2 ± 10.5 13.2 ± 8.3 #2. Ds control 120.2 ± 36.8 49.9 ± 26.8 18.1 ± 16.3 #3. linoleic acid #4. a- linolenic acid #5. y-linolenic acid 219.3 ± 93.2 97.6 ± 37.5 37.4 ± 28.5 78.9 ± 20.2 31.5 ± 4.0 36.7a 234.4 ± 115.6 144.9 ± 108.3 77.2 ± 55.4 246.7a 80.0' 62.8a 105.8a 42.8a 20.3a #6. jasmonic acid #7. antibiotic #8. arachidonic acid 199' 336.6" All numbers are ppm unit, a single run, others average of two or three runs, b sum of various HETE's including 5-,8-,11-,12-,15-HETE Table V.2 Effects on the yields of oxylipin from culture fed with six precursors experiment, a-linolenic acid might decrease production through sample toxicity to the algal cells. Jasmonic acid increased the production of three oxylipins, while addition of Ca-ionophore had no effect on the yield of oxylipins. The standard deviations in the oxylipin production observed in these studies could be due to the following reasons: firstly, biological experiments have unpredictable results because of the complexities inherent in living systems: secondly, experimental variability contributes to this deviation, including production of mass of cell density, partial oxylipin purification, oxylipin derivatization, and GC-MS analysis. #1. Ls control #2. Ds control #3. linoleic acid added #4. alpha-linolenic acid added #5. gamma-linolenic acid added #6. jasmonic acid added #7. antibiotic added i 2 3 4 5 6 7 Experiment Figure V.7 Yield of 13-HODTA from cultures added with different precursors. 400 #1. Ls control #2. Ds control 300 #3. linoleic acid added #4. alpha-linolenic acid added 200 #5. gamma-linolenic acid added #6. jasmonic acid added 100 #7. antibiotic added #8. arachidonic acid added 0 1 2 3 4 5 6 7 8 Experiment Figure V.8 Yield of 13 -NODE from cultures added with different precursors. 400 sum of various HETE's (not truely quantifiable) #1. Ls control #2. Ds control 300 #3. linoleic acid added #4. alpha-linolenic acid added 200 #5. gamma-linolenic acid added #6. jasmonic acid added 100 #7. antibiotic added #8. arachidonic acid added 0 2 3 4 5 6 7 8 Experiment Figure V.9 Yield of 15-HETE from culture added with different precursors. 0 #1. Ls control 400 #2. Ds control #3. linoleic acid added 300 E #4. alpha-linolenic acid added ca a_ #5. gamma-linolenic acid added 200 #6. jasmonic #7. antibiotic added 100 #8. arachidonic acid added 13-HODTA ra 13 -NODE 0 1 2 3 4 5 6 7 8 15-HETE Experiment Figure V.10 Comparison of the yields of three oxylipins from feeding experiment 112 EXPERIMENTAL Partial Purification of Oxylipins. Fresh wet tissue was homogenized by a Potter-Elvehjem tissue homogenizer in 20 mL of CH2C12/Me0H (2:1 v/v) (three times). Ricinelaidic acid was added to the extract as an internal standard for quantitative analysis. The extracts were pooled, filtered, and resulting solvent was dried using a Buchi rotary evaporator. A portion was then methylated with CH2N2 in Me0H at room temperature. This methylated extract was subjected to Prep-TLC using silica gel 60 F254 in 25% EtOAc /hexanes. The spot containing the desired methylated hydroxy acids was broadly scraped off the TLC plate. Several methylated oxylipins were used for references to locate the methylated hydroxy acid fraction of the extract. The resulting powdered gel was extracted with Et20, filtered, and evaporated by rotavaporation. Gas Chromatography-Mass Spectrometry (GC-MS). A portion of each oxylipin sample was derivatized to trimethylsilyl (TMS) ethers with 2 drops of hexamethyldisilazane (HMDS) and chlorotrimethylsilane (TMS-CI) in pyridine for 2 h. The reaction mixture was concentrated in vacuo and the residue was resuspended in hexanes. After centrifugation, the supernatant was injected into the GC-MS. The TMS ethers of the methyl esters of the hydroxy fatty acids were chromatographed by GC-MS using a Hewlett-Packard HP 5890 Series II gas chromatograph and HP 5971 mass spectrometer. For the injection, 14 of the sample volume was injected into a Hewlett-Packard HP-1 crosslinked methyl 113 silicone capillary column (12.5 m x 2 mm x 0.33 µm) operating with helium as the carrier gas at a temperature program of 100 °C initial, 10 °C/ min ramp, 240 °C final, 250 °C injector, and 280 °C detector. Calibration Curves and Detector Response Factors. Different stock solutions of ricinelaidic acid internal standard and each oxylipin were prepared from stock solutions in hexanes at concentrations of 1-100 g,g/j1L by serial dilution. For the injection, 14 of each was injected into the GC-MS in triplicate. Total ion peak areas of 13-HODTA and 15-HETE were measured and plotted for each concentration using Cricket graph while extract ion peak areas (m/z = 311 for 13-HODE, m/z = 187 for ricinelaidic acid) were used to resolve the problem. The detector response factors (Drf) for each oxylipin to internal standard was determined from the following relationship (1): Dry= Mis x Ao Mo x Ais (1) Muo - Mis x Auo Drf x Ais (2) where Drf is the detector response factor, Mis is the concentration of the internal standard, ricinelaidic acid, Ao is the total or extract ion peak area of oxylipin while Mo is the concentration of the oxylipin, and Ais is the the total or extract ion peak area of standard compound. Once the detector response factor of each oxylipin was obtained, the quantity of oxylipin, expressed as ppm, was calculated from the modified 114 formula (2); where, Muo is the concentration of unknown oxylipin in the sample, and Auo is the total or extract ion peak area of unknown oxylipin. 115 CHAPTER VI. PRODUCTION OF OXIDIZED ANANDAMIDE DERIVATIVES AND EVALUATION OF THEIR BINDING AFFINITY TO THE CANNABINOID RECEPTOR (CB1) ABSTRACT Five oxidized anandamide derivatives were prepared from arachidonylethanolamide (anandamide) through autooxidation, followed by reduction using triphenyiphosphine. Their structures were determined by a combination of NMR spectroscopy and GC-MS. The cannabinoid receptor binding affinity of these derivatives was evaluated. This study revealed the following trend in activity: anandamide > 15-hydroxyanandamide > 9­ hydroxyanandamide > 8-hydroxyanandamide > 11-hydroxyanandamide > 5­ hydroxyanandamide. 116 INTRODUCTION Anandamide (arachidonylethanolamide) (1) is a mammalian endogenous ligand and hydrophobic brain constituent that binds with high affinity to brain cannabinoid receptors (CB1).113 This lipid is produced and released from brain neurons' and is decomposed by amidohydrolase activity to give arachidonate and ethanolamine.115 The first discovery of this naturally occuring brain component was described by Devane et al. in 1992.11' Until the late 1980's, the misleading hypothesis was dominated that the endogenous substrate for this receptor existed as a water soluble component.' In 1990, the finding of cannabinoid receptors"' expressed at significant levels in distinct regions of the brain suggested the existence of an endogeneous ligand for this receptor. Then, in the early 1990's, alternate ideas that an endogenous cannabimimetic compound might be a lipophilic constituent like THC (A9-tetrahydrocannabinol) (2), the psychoactive principle in cannabis, resulted in the successful isolation and characterization of an endocannabinoid.111 During the past five years, researchers have revealed that anandamide shares pharmacological properties with THC and other cannabinoids in both the central nervous systems (CNS) and peripheral systems. A few structure-activity relationship (SAR) studies on anandamide have been conducted to gain insight into how anandamide and THC bind to cannabinoid receptors. These studies which would allow the development of 117 better ligands for these receptors possibly useful in therapeutical applications. Recent studies have shown that both chain length and degree of unsaturation of the fatty acid chain are critical in determining the affinity of the ligands for binding to receptors."' Hydroxylation of anandamide by lipoxygenase at different olefinic positions also resulted in changes in receptor affinity. 119,120 In this chapter, I will describe the preparation and structure elucidation of several hydroxy anandamides produced in a non-specific manner (autoxidation), followed by a discussion of the binding affinities of these derivatives to the CBI cannabinoid receptor. Ken Soderstrom in the laboratory of Dr. Thomas Murray was responsible for the competitive receptor ligand displacement assay with a variety of hydroxyanandamides. 0 H N H CH3 1 CH3 118 RESULTS AND DISCUSSION A small quantity of anandamide was oxidized (see experimental). Half of this aliquot was reduced to gain some insight into the potential activity of these derivatives on the CB1 receptor prior to large scale synthesis. Both portions of the hydroperoxy and hydroxy derivatives exibited activity in the receptor binding assay. Consequently, a large scale oxidation of anandamide was performed, followed by reduction in the presence of triphenylphosphine. Separation of these products was achieved using a Sep-Pak column containing 500 mg of silica gel, followed by normal phase HPLC. Figure VI.1 presents a general scheme for the production and the elution profile of oxidized anandamides in normal phase HPLC. Identification of each peak was determined using a combination of NMR spectroscopy and GC-EIMS spectroscopy. The peaks 2, 3, 4, 6, and 7 were identified as unreacted starting material, anandamide (1), 15­ hydroxyanandamide (3), 11-hydroxyanandamide (4), mixture of 9- and 8­ hydroxyanandamides (5) (6), and 5-hydroxyanandamide (7), respectively (Figure VI.2). GC-EIMS analysis of the methyl ester-TMS ether derivative of peak 3 identified its structure to be 15-hydroxyanandamide. Its mass spectra exhibited distinct ions at m/z 436, 335, and 173 (Figure VI. 3). The fragment ion at m/z 407 was produced from a rearrangement of the trimethylsilazane moiety and subsequent cleavege of the C14-C15 bond. This is consistent with a previous 119 0 Anandamide 1. thin film 2. 02, 5 min, rt Mixture of ROOH's 3. reduction with triphenyl phosphine 4. partial separation by Sep-Pak Mixture of hydroxy Anandamides 5. normal phase HPLC Figure VI.1 Preparation and purification of various hydroxyanandamides 120 0 1' .........,..............F.://...OH 15W) N H 11 OH 15-hydroxyanandamide (3) O NOH H HO 11-hydroxyanandamide (4) 0 9-hydroxyanandamide (5) 8-hydroxyanandamide (6) 5-hydroxyanandamide (7) Figure VI.2 Structures of various hydroxyanandamides 121 (A) Abundance 12000 Average of 18.890 to 19.086 min.: 1V90-3.D 713 173 10000 8000­ 173 6000 15-hydroxyanandamide (3) 4000 407 2000­ 260 113 50 p/z--> (B) 100 Abundance 250001 150 200 250 492 507 367 314 300 436 350 400 450 500 Average of 18.211 to 18.408 min.: 1V90-4.D 215 arms 20000 15000 11-hydroxyanandarnide (4) 100001 384 5000, 355 17116 155 204 260 29412 417 446 0 /2--> 50 100 Abundance (C) 150 200 250 300 350 400 492 507 450 500 Average of 18.231 to 18.259 min.: IV91-6B.D ( +, *) 25000 20000 15000 10000 5000 0 8/2 (D) 100 50 Abundance 150 200 250 300 350 400 450 Average of 18.853 to 18.884 min.: 1V91-6A.D (+,*) 205.­ 8000 . 7000 344 6000 8-Indronenanclanwle(8) 5000 4000 254 3000 211 2000 103 75 -> :46 281 315 I. 0 4/2 164 6 1000 SO 100 150 200 250 300 370 350 492 417 515 400 450 500 Figure V1.3 Mass spectra of (A)15-hydroxyanandamide, (B) 11­ hydroxyanandamide, (C) 9-hydroxyanandamide, (D) 8-hydroxyanandamide. 122 0 OTMS TMS A m/z = 407 OTMS m/z = 355 TMS B m/z = 355 0 NOTMS TMS C m/z = 327 0 NOTMS TMS D m/z = 315 Figure VI.4 Proposed structures of the ions at m/z 407, 355, 327, 315 formed due to rearrangement process corresponding to 15- (A), 11- (B), 9­ (C), and 8-hydroxyanandamides (D), respectively. H-5 H-6 H-8 H-9 H-11 H-17 H-18 H-19 H-2 H-20 H-1' H-2' H-4 H-7 H -3 H-12 H-10 H-14 H-13 H-15 H-16 7.0 6.5 6.0 5.5 5.0 4.5 4.0 3.5 3.0 2.5 PPM Figure VI.5 1H NMR spectrum of 15-hydroxyanandamide (3). 2.0 1.5 1.0 H -16 H-20 H-17 H 18 H-19 4.5 4.0 PPM 3.5 3.0 2.5 2.0 i.5 Figure VI.6 1H-1H COSY of 15-hydroxyanandamide (3). 4=, 125 0 5.40 5.40 2.98a 5.40 5.40 6.56 3.42 1.73 2.12 2.21 1.55 1.31 3.70 CH3 0.89 4.18 5.40 5.98 1.31 1.31 5.72 OH OH 15-hydroxyanandamide (3) Coupling constant for 3 (400 MHz, CDCI3): J2_3 = 7.4, J3-4 = 7.4, J11-12 = 11.0 = 4.7 J13-14 = 15.2, J14-15 = 6.3, J19-20 = 7.0, 0 OH 5.40 5.69 5.98 6.5 2.82a 4.20 2.98a 5.40 5.40 5.40 3.43 1.75 5.40 1.51 2.28 2.05 1.31 OH 3.72 CH3 0.89 1.31 1.31 5-hydroxyanandamide (7) Coupling constant for 7 (400 MHz, CDCI3): J2_3 = 7.4, J3.4 = 7.4, J8_8= 6.4 J6-7 = 15.2, J8.9 = 11.0, J19-20 = 7.0, J1,_2. = 4.7 Figure VI.7. 1H NMR data assignments for 15- (3) and 5-hydroxyanandamides (7) alnterchangeable. H-20 H-17 H-18 H-19 H-9 H-11 H-2 H-2' H-12 H-14 H-15 H-4 H-1' H-13 H-16 H-10 H-3 H-8 H-7 H-5 H-6 It\ 7.0 6.5 6.0 5.5 5.0 4.5 4,0 PPM 3.5 3.0 2.5 2.0 Figure VI.8 1H NMR spectrum of 5-hydroxyanandamide (7). 15 1.0 H-2 H-3 H-4 H-5 cmo O 03 a O 7.0 6.5 6.0 5.5 5.0 4.5 4.0 PPM 3.5 3.0 2.5 2.0 Figure VI.9 11-1-1I-1 COSY of 5-hydroxyanandamide (7). 1.5 1.0 128 report (Figure VI.4). 119,120 Additional analysis of the one- and two-dimensional NMR spectra of this compound confirmed it as 15-hydroxyanandamide (3) (Figure VI.5). The 1H NMR spectrum showed the presence of one 0-bearing methine resonance at 5 4.18 and conjugated olefinic proton resonances at S 6.56, 5 5.98, 5 5.72, and 5 5.40. The presence of two methylene proton resonances at 5 2.82 and at 5 2.98 in the 1H NMR spectrum of this product suggested that it was possibly either 5- or 15-hydroxyanandamide. Other possible derivatives, including 8-, 9-, 11-, and 12-hydroxyanandamides, could be eliminated because these derivatives do not have two methylene proton resonances but rather contain a single proton resonance around 5 2.82 to S 2.98. Further analysis of the 1H-1H COSY spectrum indicated the identity of peak 3 as 15-hydroxyanandamide (3) (Figure VI.6). The methine proton resonance at S 4.18 bearing the hydroxyl group showed COSY correlation to the methylene proton resonance at 8 1.55 (H-16) which was correlated to three methylene proton resonances at 8 1.31 (H-17, H-18, H-19). Furthermore, the 1H NMR spectrum of this compound was compared to that of 15­ hydroxyanandamide previously reported and it proved to be identical.119 The 1H NMR data assignments for 15- (3) and 5-hydroxyanandamides (7) are shown in Figure VI.7. Peak 4 was identified as 11-hydroxyanandamide (4) on the basis of GC­ EIMS, observing significant fragment patterns at m/z 384 and 225. A fragment 129 ion peak at m/z 355 was also produced from a rearrangement of 11­ hydroxyanandamide (4). Peak 6 was found to be mixture of the 9-and 8-hydroxyanandamides as determined by 11-I NMR spectroscopy. Repeated normal phase HPLC of this fraction eventually yielded separation of the two derivatives. The first portion was identified as 9-hydroxyanandamide (5). The identity of this product was supported by GC-EIMS, exhibiting ions at m/z 356 together with 327 as a fragment ion from a rearrangement process. The second portion was verified as 8-hydroxyanandamide (6) which was characterized by ions at m/z 344 and 265. The peak observed at m/z 315 was due to a rearrangement of 6. Peak 7 was identified as 5-hydroxyanandamide (7) based on detailed analysis by 1H NMR spectroscopy (Figure VI.8). Analysis of the 1H NMR suggested either 5- or 15-hydroxyanandamide based on the presence of the two methylene proton resonances at 6 2.97 and 6 2.85. The 1H NMR spectrum of this peak showed the presence of a methine (8 4.2, H-5) bearing a secondary hydroxyl group whose location, 5-hydroxy, was shown by spin couplings in 1H­ 1H COSY spectrum (Figure VI.9). In the 1H-1H COSY, the methine proton resonance at 8 4.2 bearing the hydroxyl group showed a correlation to the methylene proton resonance at 8 1.51 (H-4) which was correlated to another methylene proton resonance at 6 1.75 (H-3). This methylene at 8 1.75 (H-3) was further correlated to the methylene protons at 6 2.28 (H-2) proximate to the amide ketone group. 130 The Yamamoto group in 1995 reported inhibitory effects on electricallyevoked contraction of mouse vas deferens assay with 12S- and 15S­ hydroxyanandamides prepared from the incubation of [14C]anandamide with several purified lipoxygenases.119 The 15-hydroxyanandamide was active with an IC50 of 0.63 !AM as well as anandamide (0.17 gM), while the 12­ hydroxyanandamide was inactive ( > 10 liM). However, the cannabinoid binding assay was not performed with these derivatives. At the same time, the Bornheim group reported a CB1-binding assay with 11-, 12-, and 15-hydroxyanandamides.12° The 12-hydroxyanandamide was the most active with Ki (affinity of ligand for receptor) of 0.031 p,M. Anandamide was the next most active with Ki of 0.071 liM, followed by the 15­ hydroxyanandamide (0.418 p,M), and 11-hydroxyanandamide (1.1 p,M). In this study, we explored the effect of hydroxylation of anandamide on the activity in the CB1 receptor binding assay. Anandamide was the most active with Ki of 0.51 µM in this assay (Figure VI.9). The fact that 15­ hydroxyanandamide (3) was active (0.74 p,M) while 11-hydroxyanandamide (4) was inactive (3.3 p,M) is in good agreement with the results described above. The significance of this study is the discovery that the 9- (5) and 8­ hydroxyanandamide (6) are active at 2.6 pM and 2.1 gM, respectively. The 5­ hydroxyanandamide (7), a new compound, was found to be relatively inactive (8.24 p.M). All of these hydroxy derivatives are racemic. Further study is 100 90 80 70 60 50 40 30 20 Anandamide Ki = 0.51 RM 0 15-Hydroxyanandamide KI = 0.74 piM Mixture of 9- and 8-Hydroxyanandamide Ki = 1.83 WI 11-Hydroxyanandamide Ki = 3.30IAM A 5-Hydroxyanandamide Ki = 8.24 p.M 10 0 -8 1 -7 -6 1 -5 Iog(Competitor) I -4 , -3 Figure VI.10 Inhibition of the specific binding of [3H]CP -55940 to rat neuronal membrane by anandamide and oxidized anandamide derivatives. Specific Binding 132 ongoing to attempt chiral separation of these mixtures, and to investigate which form of the isomers is active to CB1 receptor binding. EXPERIMENTAL General Experimental Procedures. NMR spectra were recorded on a Bruker AM 400 spectrometer in the solvent specified. Chemical shifts were referenced to solvent CDCI3 signal at 7.27 ppm for 1H NMR. Gas chromatography/mass spectrometry (GC/MS) was carried out utilizing a Hewlett-Packard 5890 Series II GC connected to a Hewlett-Packard 5971 mass spectrometer. Production and Isolation of Hydroxyanandamides. Anandamide (8 mg) purchased from Cayman Chemicals (Ann Arbor, MI) was added into a round bottom flask (250 mL). Pure molecular oxygen was charged for five minutes into the flask which was slowly rotated on a rotary evaporater to make a thin film on the surface of the flask. The reaction was terminated after 4 hr. The resulting hydroperoxide derivatives were reduced with triphenylphosphine (5.0 mg) in 2.0 mL Et20, and the reaction mixture was stirred at rt for 2 hr. The solvent was reduced by a rotary evaporater. The concentrate was diluted with 50% EtOAc /hexanes, and passed through a Si Sep-Pak column containing 500 mg of silica gel to remove triphenylphosphine. The desired products were further eluted by 10% Me0H/CHCI3 on Sep-Pak. Repetitive HPLC on a Phenomenex 133 Si column (10 mm, 500 x 10 mm) of the oxidized products at 9 mL/min with 65:32:3 (v/v) EtOAc /hexanes /MeOH resulted in the elution of 15­ hydroxyanandamide (0.5 mg), 11-hydroxyanandamide (0.2 mg), 9­ hydroxyanandamide (0.2 mg), 8-hydroxyanandamide (0.2 mg), and 5­ hydroxyanandamide (0.4 mg), respectively. Gas Chromatography-Mass Spectrometry (GC-MS). A portion of oxidized anandamide derivatives was derivatized to trimethylsilyl (TMS) ethers with 2 drops of hexamethyldisilazane (HMDS), 2 drops chlorotrimethylsilane (TMS-CI), and 2 drops pyridine for 2 h. The reaction mixture was concentrated in vacuo and the residue was resuspended in hexanes. After centrifugation, the supernatant was injected into the GC-MS. The TMS ethers of the hydroxyanandamides were chromatographed by GC-MS using a HewlettPackard HP 5890 Series II gas chromatograph and HP 5971 mass spectrometer. For the injection, li_tl_ of the sample volume was injected into a Hewlett-Packard HP-1 crosslinked methyl silicone capillary column (12.5 m x 2 mm x 0.33 p.m) operating with helium as the carrier gas at a temperature program of 100 °C initial, 10 °C/ min ramp, 240 °C final, 250 °C injector, and 280 °C detector. Evaluation of hydroxyanandamides as ligands to the cannabinoid receptor Ken Soderman in the laboratory of Dr. Thomas Murray was 134 responsible for the competitive receptor ligand displacement assay with a variety of hydroxyanandamides. 135 CHAPTER VII CONCLUSION Until recently marine cyanobacteria have not been explored extensively for their potential uses as pharmaceuticals. However, they are emerging as a new source of novel biologically active metabolites. Particulary, various collections of L. majuscula have provided diverse bioactive secondary metabolites. In this regard, my interest has focused on the investigation of L. majuscula. As a part of our research project aimed at the discovery of new bioactive compounds from marine algae, we isolated two new and potent antimitotic metabolites, curacins B and C, from a Curacao collection of Lyngbya majuscula. In addition, four curacin A analogs were prepared by semisynthetic efforts. The biological and biochemical properties of the new natural products and synthetic derivatives of curacin A were examined. The isolation and identification of curacins from L. majuscula prompted us to investigate another collection of this species. Consequently, a new cytotoxic lipopeptide, microcolin C along with three known compounds, microcolins A, B, and D, were isolated by bioassay-guided fractionation of the organic extract from Curacao and Granada collections of Lyngbya majuscula. Microcolin C was found to have an interesting profile of cytotoxicity to human cancer-derived cell lines. Microcolins A-C also exhibited potent toxicity to brine shrimp (LC50 of 0.004 µg /ml, 0.4 µg /ml, and 0.2 µg /ml, respectively). 136 Investigation of an Indonesian red alga Vidalia sp. yielded a new secondary metabolite, vidalenolone. The structure of this new cyclopentenolone-containing compound was determined by a combination of spectroscopic methods. Filamentous cells isolated from female gametophytes of the brown alga Laminaria saccharina were cultured in a flask or bioreactor culture. These cultures produced a variety of 15-lipoxygenase metabolites. Feeding experiments were carried out to attempt the enhancement of these 15­ lipoxygenase metabolites. The presence of linoleic acid, y-linolenic acid and jasmonic acid increased the production of the three oxylipins, 13-hydroxy-9,11­ octadecadienoic acid (13-HODE), 13-hydroxy-6,9,11,15-octadecatetraenoic acid (13-HODTA), and 15-hydroxy-5,8,11,13-eicosatetraenoic acid (15-HETE), 2 to 4 times over the controls. On the other hand, addition of antibiotic did not increase the yield, while a-linolenic acid suppressed production of these metabolites. Arachidonic acid addition gave rise to a variety of HETE products via autooxidation. I described the preparation and structure elucidation of several hydroxy anandamides including 5-, 8-, 9-, 11-, and 15-hydroxyanandamide. The aim of this work was to develop better ligands for these receptors which may be of some use in therapeutical applications. This study on the cannabinoid receptor binding affinity revealed the following trend in activity: anandamide > 15­ hydroxyanandamide > 9-hydroxyanandamide > 8-hydroxyanandamide > 11­ hydroxyanandamide > 5-hydroxyanandamide. 137 In conclusion, the overall hypothesis of this work is based on the published literature that marine organisms, especially marine algae, have been an important source of structurally novel classes of bioactive compounds. Hence, continued investigations of these life forms for this purpose seems reasonable and warrented. My investigations reported in this thesis of various marine algae have yielded several new and bioactive metabolites, such as curacins B and C, microcolin C, and vidalenolone. Thus, these findings indicate that marine algae are a reliable source for bioactive metabolites and there is more work to be investigated with these marine organisms as a source of potential drugs. 138 BIBLIOGRAPHY 1. Farnsworth, N. R. Screening plants for new medicines. In: Biodiversity (Eds. Wilson, E. 0. and Peters, F. M.), Academic Press, New York, 1988, 61-73. 2. Barth, R. H.; Broshears, R. E. The invertebrate world, CBS College Publishing, New York, New York. 1982, 646. 3. Bergmann, W. J. Mar. Res. 1949, 8, 137-176. 4. Carte, B. K. Bio Science 1996, 46, 271-286. 5. Clark, A. M. Pharmaceutical Research, 1996, 13, 1133-1141. 6. Fenical, W. Chem. Rev. 1993, 93, 1673-1683. 7. 8. Burkholder, P. R.; Pfister, R. M.; Leitz, F. P. Appl. Microbiol. 1966, 14, 649. Laatsch, H.; Pudleiner, H. Liebigs. Ann. Chem. 1989, 9, 863. 9. Needham, J.; Andersen, R.; Kelly, M. T. Tetrahedron Lett. 1991, 32, 315. 10. Gustafson, K.; Roman, M.; Fenical, W. J. Am. Chem. Sco. 1989, 111, 7519-7524. 11. Tapiolas, D. M.; Roman, M.; Fenical, W. J. Am. Chem. Soc. 1991, 113, 4682-4683. 12. Trischman, J. A.; Tapiolas, D. M.; Jensen, P. R.; Dwight, R.; Fenical, F. J. Am. Chem. Soc. 1994, / /6, 757-758. 13. Kobayashi, J.; Ishibashi, M. The Alkaloids , Brossi, A.; Cordell, G. A. Academic Press: San Diego, 1992, 41, 41-124. 14. Yasumoto, T; Yasumura, D.; Yotsu, M.; Endo, A.; Kotaki, Y. Agric. Bio. Chem. 1986, 50, 793-795. 15. Do, H. K.; Kogure, K.; Simidu, U. Applied and Environmental Microbiology, 1990, 56, 1162-1163. 16. Ikawa, M.; Wegner, K.; Faxall, T. L.; Sasner, J. J., Jr. Toxicon 1982, 20, 747. 17. Kodama, M.; Ogata, T.; Sato, S. Agric. Biol. Chem. 1988, 52, 1075. 139 18. Moore, R. E. Constituents of blue-green algae in Marine natural products: Chemical and biological perspectives, Vol. IV. P. J. Scheuer, ed. Academic Press, New York. 1981, 1-52. 19. Francis, G. Nature 1878, 18, 11-12. 20. Bishop, C. T.; Anet, E.; Gorham, P. R. Can. J. Biochem. Physiol. 1959, 37, 453-471. 21. Carmichael, W. W.; Beasley, V.; Bunner, D. L.; Eloff, J. N.; Falconer, (.;Gorham, P.; Harada, K.; Krishnamurthy, T.; Min-Juan, Y.; Moore, R. E.; Rinehart, K.; Runnegar, M.; Skulberg, 0. M.; Watanabe, M. Toxicon 1988, 26, 236-241. 22. Carmichae, W. W. J. Appl. Bacteriol. 1992, 72, 445-449. 23. Falconer, I. R.; Beresford, A. M.; Runnegar, M. T. C. Med. J. Chem. 1983, 1, 511-514. 24. Rinehart, K.; Harada, K.; Namikoshi, M.; Chen, C.; Harvis, C. A. J. Am. Chem. Sco. 1988, 110, 8557-8558. 25. Ohta, T.; Sueoka, E.; lida, N.; Komori, A.; Suganuma, M.; Nishiwaki, R.; Tatematsu, M.; Kim, S-J.; Carmichael, W. W.; Fujiki, H. Cancer Res. 1994, 54, 6402-6406. 26. Stratmann, K.; Burgoyne, L.; Moore, R. E.; Patterson, G. M. J. Org. chem. 1994, 59, 7219-7226. 27. Trimurtulu, G.; Ohtani, I.; Patterson, G. M.; Moore, R. E.; Corbett, T. H.; Valeriote, F. A.; Demchik, L. J. Am. Chem. Soc. 1994, 116, 4729-4737. 28. Trimurtulu, G.; Ogino, J.; Heltzel, C. E.; Husebo, T. L.; Jensen, C. M.; Larsen, L. K.; Patterson, G. M.; Moore, R. E.; Mooberry, S. L.; Corbett, T. H.; Valeriote, F. A.J. Am. Chem. Soc. 1995, 117, 12030-12049. 29. Smith, C. D.; Zhang, X. J. Biol. Chem. 1996, 271, 6192-6198. 30. Cardellina, J. H.; Marner, F-J.; Moore, R. E. Science, 1979, 204, 193­ 195. 31. Carter, D. C.; Moore, R. E.; Mynderse, J. S.; Niemczura, W. P.; Todd, J. S. J. Org. Chem. 1984, 49, 236-241. 140 32. Gerwick, W. H.; Jiang, J. D.; Agarwal, S. K.; Farmer, B. Tetrahedron, 1992, 48, 2313-2324. 33. Frankmolle, W. P.; Knubel. G.; Moore, R. E.; Patterson, G. M. L. J. Antibiotics, 1992, 45, 1458-1466. 34. Mayer, A. M. S.; Jacobs, R. Manoalide: An antiinflammatory and analgesic marine natural product In Biomedical importance of marine organisms, (Fautin, D.) California Academy of Sciences, San francisco, 1988, 133-142. 35. Campbell, W. B.; Halushka, P. V. Lipid-Derived Autocoids: Eicosanoids and Platelet-Activating Factor In The Pharmacological Basis of Therapeutics, 9th ed.; Hardman, J. G.; limbird, L. E.; Molinoff, P. B.; Ruddon, R. W.; Gilman, A. G., Ed; Mcgraw-Hill: New York, 1996, 601­ 616. 36. Cohen, M. M. Biological protection with prostaglandins, Vol. I. CRC Press, Inc., Boca Raton, FL. 1985. 37. Feuerstein, G.; Hallenbeck, J. M. FASEB, 1987, 1, 186-192. 38. Piomelli, D. Arachidonic Acid in Cell Signalling, Champman & Hall, 1996, 15-53. 39. Weinheimer, A. J.; Sparggins, R. L. Tetrahedron Lett. 1969, 59, 5185­ 5188. 40. Coll, J. C. Chem. Rev. 1992, 92, 613-631. 41. Carmely, S.; Kashman, Y.; Loya, Y.; Benayahu, Y. Tetrahedron Lett. 1980, 21, 875-878. 42. Gerwick, W. H.; Moghaddam, M.; Hamberg, M. Archives of Biochemistry and Biophysics, 1991, 290, 436-444. 43. Gregson, R. P.; Marwood, J. F.; Quinn, R. J. Tetrahedron Lett. 1979, 46, 4505-4506. 44. Gerwick, W. H.; Nagle, D. G.; Proteau, P. J. Topics in Current Chemistry, Marine Natural Products-Diversaty and Biosynthesis, (Scheuer, P. J. Ed), Springer-Verlag Berlin Heidelberg, 1993, Vol. 167, 117-180. 45. Gerwick, W. H. Chem. Rev. 1993, 93, 1807-1823. 141 46. Gerwick. W. H.; Bernart, M. W. in Pharmaceutical & Bioactive Natural Products. Marine Biotechnology, Vol 1 (Attaway, D. W.; Zaborsky, 0. R. Eds), Plenum Press, New York,1993, 101-152. 47. Gerwick, W. H. Biochimica et Biophysica Acta, 1994, 1211, 243-255. 48. Gerwick, W. H. Lipid, 1996, 31, 1215-1230. 49. Cardellina Ii, J. H.; Dalietos, D.; Marner, F-J.; Mynderse, J. S.; Moore, R. E. Phytochemistry, 1978, 17, 2091-2095. 50. Cardellina II, J. H.; Moore, R. E. Tetrahedron, 1980, 36, 993-996. 51. Nagle, D. G.; Gerwick. W. H. Tetrahedron Lett. 1990, 31, 2995-2998. 52. Solem, M. L.; Jiang, Z. D.; Gerwick, W. H. Lipids, 1989, 24, 256-260. 53. Kugel, C.; Lellouche, J-P.; Beaucourt, J.-P. Tetrahedron Lett. 1989, 30, 4947-4959. 54. Gundlach, H.; Muller, M.; Kutchan, T. M.; Zenk, M. H. Proc. Natl, Acad. Sci. USA, 1992, 89, 2389-2393. 55. Pollio, A.; Greca, M. D.; Monaco, P.; Pinto, G.; Previtera, L. Biochim. Biophys. Acta 1987, 963, 53-60. 56. Bernart, M. W.; Gerwick. W. H. J. Nat. Prod. 1992, 56, 245-259. 57. Proteau, P. J.; Gerwick, W. H. Tetrahedron Lett. 1992, 33, 4393-4396. 58. Gardner, H. W. Biochim. Biophys. Acta 1991, 1084, 221-239. 59. Todd, J. S.; Proteau, P. J.; Gerwick, W. H. Tetrahedron Lett. 1993, 34, 7689-7692. 60. Kato, Y; Scheuer, P. J. Pure Appl. Chem. 1975, 41, 1. 61. Cardellina, J. H.; Marner, F.-J.; Moore, R. E. Science, 1979, 204, 193. 62. Cardellina, J. H.; Moore, R. E. Tetrahedron Lett., 1979, 22, 2007-2010. 63. Simmons, C. J.; Marner, F.-J.; Cardellina, J. H.; Moore, R. E.; Seff, K. Tetrahedron Lett., 1979, 22, 2003-2006. 64. Orjala, J.; Nagle, D. G.; Hsu, V. L.; Gerwick, W. H. J. Am. Chem. Soc. 1995, 117, 8281-8282. 142 65. Wu, M., Ph.D. Thesis, Oregon State University, Corvallis, 1996, 18-47. 66. Gerwick, W.H.; Proteau, P.J. ; Nagle, D.G.; Hamel, E. ; Blokhin, A.; Slate, D.L. J. Org. Chem. , 1994, 59, 1243-1245. 67. Hamel, E.; Blokhin, a. V.; Nagle, D. G.; Yoo, H. D.; Gerwick, W. H. Drug Dev. Res. 1995, 34, 110-120. 68. Yoo, H. D.; Gerwick, W. H. J. Nat. Prod. 1995, 58, 1961-1965. 69. Couperus, P. A.; Clague, A. D. H.; Dongen, J. P. C. M. Org. Magn. Reson. 1976, 8, 426-431. 70. Nagle, D. G.; Gerald, G. S.; Yoo, H. D.; Gerwick, W. H.; Kim,T. S.; Nambu, M.; White, J. D.Tetrahedron Lett., 1995, 36, 1189-1192. 71. Blokhin, A. V.; Yoo, H. D.; Geralds, R. S.;Nagle, D. G.; Hamel, E.; Gerwick, W. H. Molecular pharmacology 1995, 48, 523-531. 72. Takahashi, M.; Iwasaki, S.; Kobayashi, H.; Okuda, S.; Murai, T.; Sato, Y. Biochim. Biophys. Acta 1987, 929, 215-223. 73. Pettit, G. R.; Kamano, Y.; Herald, C. L.; Tuinman, A. A.; Boettner, F. E.; Kizu, H.; Schmidt, J. M.; Baczynskyj, L.; Tomer, K. B.; Bontems, R. J. J. Am. Chem. Soc. 1987, 109, 6883-6885. 74. Bollag, D. M.; McQueney, P. A.; Zhu, J.; Hensens, 0; Koupal, L.; Liesch, J.; Goetz, M.; Lazarides, E.; Woods, C. M. Cancer Res., 1995, 55, 2325­ 2333. 75. Pinard, P. V.; Lai, J. Y.; Yoo, H. D.; Nagle, D. G.; Marquez, B.; Falck, J. R.; Gerwick, W. H.; Hamel, E.; Nambu, M; White, J. D. Molecular Pharmacology in press 1997. 76. Moore, R.E.; Patterson, G.ML.; Carmichael, W.W. Memoirs Calif. Acad. Sci, No. 13, San Francisco, 1988, pp 143-150; Carmichael, W.W., J. Appl. Bact., 1992,72, 445-459. 77. Marner, F.-J.; Moore, R. E. Phytochemistry, 1978, 17, 553-554 78. Cardellina, J. H.; Marner, F.-J.; Moore, R. E. J. Am. Chem. Sco. 1979, 101, 240. 79. Nagle, D. G.; Paul, V. J.; Robert, M. A. Tetrahedron Lett. 1996, 37, 6263­ 6266. 143 80. Petcher, T. Y.; Weber, H. P.; Ruegger, A. He Iv. Chim. Acta. 1976, 59, 1480- 1489. 81. Tanaka, H.; Kuroda, A.; Marusawa, H.; Hatanaka, H.; Kino, T.; Goto, T.; Hashimoto, M. J. Am. Chem. Sco. 1987, 109, 5031-5033. 82. Vezina, C.; Kudelski, A.; Sehgal, S. N. J. Antibiotics, 1975, 28, 721-726. 83. Kay, J. E.; Benzie, C. R.; Goodier, M. R.; Wick, C. J.; Doe, S. E. A. Immunology 1989, 67, 473-477. 84. Kay, J. E.; Moore, A. L.; Doe, S. E. A.; Benzie, C. R.; Schonbrunner, R.; Schmid, F. X.; Halestrap, A. P. Transplantation Proc. 1990, 22, 96-99. 85. Koehn, F. E.; Long ley, R. E.; Reed, J. K. J. Nat. Prod., 1992, 55, 613-619. 86. Koehn, F. E.; 0. J. McConnell, 0. J.; Long ley, R. E.; Sennett, S. H.; Reed, J. K. J. Med. Chem., 1994, 37, 3181-3186. 87. Mattern, R. -H.; Gunasekera, S.; McConnell, 0. Tetrahedron, 1996,52, 425-434. 88. Decicco, C. P.; Grover, P. J. Org. Chem., 1996,61, 3534-3541. 89. Andrus, M. B., Li, W.; Keyes, R. F. J. Org. Chem., 1997, 62, 5542-5549. 90. Laumen, K; Schneider, M. Tetrahedron Lett., 1985, 26, 2073-2076. 91. Deardorff, D.; Mathews, A. J.; McMekin, D. S.; Craney, C. L. Tetrahedron Lett., 1986, 27, 1255-1256. 92. Herzig, J. J. Org. Chem., 1986, 51, 727-730. 93. Boyd, M. R.; Paull, K. D. Drug Development research, 1995, 34, 91-109. 94. Hoek, C. V. D.; Mann, D. G.; Jahns, H. M. "Algae, an introduction to phycology" Cambridge University Press, 1995, 48-95 95. Stallard, M. 0.; Faulkner, D. J. Chemical Constituents of the Digestive Glandin of the Sea Hare Aplysia californica-I. Importance of Diet.Comp. Biochem. Physiol. B, 1974, 49, 25-35. 96. Fuller, R. W.; Cardellina, J. H.; Jurek, J.; Scheuer, P.; Lindner, B. A.; McGuire, M.; Gary, G. N.; Steiner, J. R.; Clardy, J.; Menez, E.; Shoemaker, R. H.; Newman, D. J.; Snader, K. M.; Boyd, M. R. J. Med. Chem. 1994, 37, 4407-4411. 144 97. Wiemer, D. F.; Idler, D. D.; Fenical, W. Experientia, 1991, 47, 851-853. 98. Patti, A.; Morrone, R. J. Nat. Prod. 1992, 1, 53-57. 99. Kazlauskas, R.; Murphy, P. T.; Wells, R. J. Aust. J. Chem. 1982, 35, 219­ 220. 100. Thiericke, R.; Zeeck, A.; Nakagawa, A.; Omura, S.; Herrold, R. E.; Wu, S. T. S.; Beale, J. M.; Floss, H. J. Am. Chem. Soc. 1990, 112, 3979-3987. 101. Clissold, D; Thickitt, C. Natural Product Report, 1994, 611-637. 102. Cragg, G. M.; Schepartz, S. A.; Suffness, M.; Greyer, M. R. J. Nat. Prod. 1993, 56, 1657-1668. 103. Rouhi, A. M. C&EN 1995, 42-44. 104. Anderson, I New Scientist 1995, 2005, 5. 105. Pettit, G. R.; Kamano, Y.; Herald, C. L.; Tuinman, A. A.; Boettner, F. E.; Kizu, H.; Tomer, K. B.; Bonterms, R. J. J. Am. Chem. Soc. 1987, 109, 6883-6885. 106. White, J. D.; Kim, T. S.; Nambu, M. J. Am. Chem. Soc. 1995, 117, 5612­ 5613. 107. Rossi, J. V.; Roberts, M. A.; Yoo, H.-D.; Gerwick. W. H. J. Appl. Phycol. 1997, 0, 1-10. 108. Hirosawa, T.; Wolk, P. J. Gen. Microbiol. 1979, 114, 433-441. 109. Armstrong, J. F.; Janda, K. E.; Alvarado, B.; Wright, A. E. J. Appl. Phycol. 1991,3, 227-282. 110. Panda, A. K.; Mishra, S.; Bisaria, V. S.; Bhojwani, S. S. Enzyme and Microbial technology, 1989, 11, 386-397. 111. Rorrer, G. L.; Modrell, J.; Zhi, C.; Yoo, H.-D.; Nagle, D. G.; Gerwick, W. H. J. Appl. Phycol. 1995, 7, 187-198. 112. Personal communication with Dr. W. H. Gerwick. 113. Devane, D. A.; Hanus, L.; Breuer, A.; Pertwee, R. G.; Stevenson, L. A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; mechoulam, R. Science, 1992, 258, 1946-1949. 114. Marzo, V. D.; Fontana, A.; cadas, H.; Schinelli, S.; Cimino, G.; Schwartz, J. C.; Piomelli, D. Nature, 1994, 372, 686-691. 145 115. Desarnaud, F.; Cadas, H.; Piomelli, D. J. Biol. Chem., 1995, 270, 6030­ 6035. 116. Evans, D. M.; Lake, J. T.; Johnson, M. R.; Howlett, A. C. J. Pharmacol Exper. Ther. 1994, 268, 1271-1277. 117. Matsuda, L. A.; Lolait, S. J.; Brownstein, M. J.; Young, A. C.; Bonner, T. I. Nature, 1990, 346, 561-564 118. Felder, C. C.; Briley, E. M.; Axelrod, J.; Simpson, J. T.; Mackie, K.; Devane, W. A. Proc. Natl. Acad. Sc!. USA 1993, 90, 7656-7660. 119. Ueda, N.; Yamamoto, K.; Yamamoto, S.; Tokunaga, T.; Shirakawa, E.; Shinkai, H.; Ofawa, M.; Sato, T.; Kudo, I.; Inoue, K,; Takizawa, H.; Nagano, T.; Hirobe, M.; Matsuki, N.; Saito, H. Biochimica et Biophysica Acta 1995, 1254, 127-134. 120. Hampson, A.; Hill, W. A. G.; Zan-Phillips, M.; Makriyannis, A.; Leung, E.; Eglen, R. M.; Bornheim, L. M. Biochimica et Biophysica Acta 1995, 1259, 173-179. 146 APPENDIX Patellamide C I .1 . 8.0 7.5 7.0 6.5 i 6.0 5.5 5.0 4.5 4.0 PPM 3.5 3.0 Figure A.1 11-1 NMR spectrum of patellamide C in CDCI3. 180 170 160 150 140 130 120 110 go 9Q PPM 80 70 60 50 40 Figure A.2 13C NMR spectrum of patellamide C in CDCI3. 30 20 10 0