This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this
material constitutes acceptance of that license and the conditions of use of materials on this site.
Copyright 2006, The Johns Hopkins University and Karl W. Broman. All rights reserved. Use of these materials
permitted only in accordance with license rights granted. Materials provided “AS IS”; no representations or
warranties provided. User assumes all responsibility for use, and all liability related thereto, and must independently
review all materials for accuracy and efficacy. May contain materials owned by others. User is responsible for
obtaining permissions for use from third parties as needed.
Underlying group dist’ns
Standard ANOVA model
µ8
Data
µ7
µ6
µ5
µ4
µ3
µ2
µ1
The ANOVA model
Three different ways to describe the model:
A.
Yti independent with Yti ∼ N(µt, σ 2)
B.
Yti = µt + ti where ti ∼ iid N(0, σ 2)
C.
Yti = µ + τt + ti where ti ∼ iid N(0, σ 2) and
P
t τt
=0
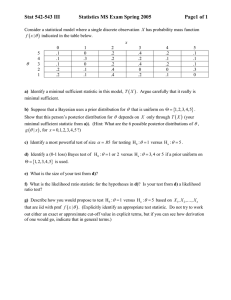
Example
For each of 8 mothers and 8 fathers, we observe (estimates of)
the number of crossovers, genome-wide, in a set of independent
meiotic products.
Question:
Do the fathers (or mothers) vary in the number of
crossovers they deliver?
Female meioses
1416
1413
Family
1362
1347
1332
1331
884
102
25
30
35
40
45
Total no. crossovers
50
55
60
Male meioses
1416
1413
Family
1362
1347
1332
1331
884
102
15
20
25
30
Total no. crossovers
ANOVA tables
Female meioses:
source
between families
within families
total
SS df
MS
F P-value
1485 7 212.2 4.60 0.0002
3873 84 46.1
5358 91
Male meioses:
source
between families
within families
total
SS df MS
F P-value
114 7 16.3 1.23
0.30
1112 84 13.2
1226 91
Permutation test
The P-values calculated above rely on the assumption that the
measurements in the underlying populations are normally distributed.
Alternatively, one may use a permutation (aka randomization) test
to obtain P-values.
1. Permute (shuffle) the XO counts relative to the family IDs.
2. Re-calculate the F statistic.
3. Repeat (1) and (2) many times (e.g., 1000 or 10,000 times).
4. Estimate the P-value as the proportion of the F statistics
from permuted data that are ≥ the observed F statistic.
Permutation dist’n : Females
^
P=0
0
1
2
3
F statistic
4
5
Observed
Permutation dist’n : Males
^
P = 32%
0
1
2
Observed
3
4
5
F statistic
Another example
A
B
200
400
600
800
1000
1200
1400
1600
1800
2000
treatment response
Are the population means the same?
By now, we know two ways of testing that:
• two-sample t-test
• ANOVA with two treatments
But do they give similar results?
ANOVA table
source
sum of squares
between treatments
SB =
X
SW =
XX
t
within treatments
ST =
i
XX
t
mean square
k–1
MB = SB/(k – 1)
(Yti − Ȳt·)2
N–k
MW = SW/(N – k)
(Yti − Ȳ··)2
(N – 1)
nt(Ȳt· − Ȳ··)2
t
total
df
i
ANOVA for two groups
The ANOVA test statistic is
MB/MW,
with
MB = n1(Ȳ1 − Ȳ··)2 + n2(Ȳ2 − Ȳ··)2
and
MW =
Pn1
Pn2
2
2
(Y
−
Ȳ
)
+
1
1i
i=1
i = 1 (Y2i − Ȳ2)
n1 + n2 − 2
Two-sample t-test
The test statistic for the two sample t-test is
Ȳ1 − Ȳ2
t= p
s 1/n1 + 1/n2
with
s2 =
Pn2
2
2
(Y
−
Ȳ
)
+
1
1i
i = 1 (Y2i − Ȳ2)
i=1
n1 + n2 − 2
Pn1
This also assumes equal variance within the groups!
Result
MB
= t2
MW
Reference distributions
If there was no difference in means, then
MB
∼ F1,n1+n2−2
MW
t ∼ tn1+n2−2
Now does this mean
F1,n1+n2−2 = (tn1+n2−2)2
?
A few facts
F1,k = t2k
Fk,∞ =
χ2
k
k
N(0,1)2 = χ21 = F1,∞ = t2∞
F table
1
2
1
·
2
·
·
·
t2k2
k2
·
k1
·
·
∞
·
·
·
Fk1,k2
·
·
·
·
·
·
∞
t2∞
·
·
·
χ2k1
k1
Underlying group dist’ns
Standard ANOVA model
µ8
Data
µ7
µ6
µ5
µ4
µ3
µ2
µ1
Underlying group dist’ns
Dist’n of group means
Random effects
model
µ8
µ
Data
µ7
µ6
Observed underlying
group means
µ5
µ4
µ3
µ2
µ1
The random effects model
Two different ways to describe the model:
A.
µt ∼ iid N(µ, σA2 )
Yti = µt + ti where ti ∼ iid N(0, σ 2)
B.
τt ∼ iid N(0, σA2 )
Yti = µ + τt + ti where ti ∼ iid N(0, σ 2)
−→
We add another layer of sampling.
Hypothesis testing
In the standard ANOVA model, we considered the µt as fixed but
unknown quantities.
We test the hypothesis H0 : µ1 = · · · = µk using the statistics
MB/MW from the ANOVA table and the comparing this to an F(k –
1, N – k) distribution.
In the random effects model, we consider the µt as random draws
from a normal distribution with mean µ and variance σA2 .
We seek to test the hypothesis H0 : σA2 = 0 versus Ha : σA2 > 0.
As it turns out, we end up with the same test statistic and same
null distribution.
Estimation
For the random effects model it can be shown that
E(MB) = σ 2 + n0 × σA2
where
P 2
n
1
N − Pt t
n0 =
k–1
t nt
Recall also that E(MW) = σ 2.
Thus, we may estimate σ 2 by σ̂ 2 = MW.
And we may estimate σA2 by σ̂A2 = (MB − MW)/n0
(provided that this is ≥ 0).
The first example
The samples sizes for the 8 families were (14, 12, 11, 10, 10, 11, 15, 9), for a total
sample size of 92.
Thus, n0 ≈ 11.45.
For the female meioses, MB = 212 and MW = 46. Thus
√
σ̂ = 46 = 6.8
(Note: overall sample mean = 40.3)
p
σ̂A = (212 − 46)/11.45 = 3.81.
For the male meioses, MB = 16.3 and MW = 13.2. Thus
√
(Note: overall sample mean = 22.8)
σ̂ = 13.2 = 3.6
p
σ̂A = (16.3 − 13.2)/11.45 = 0.52.