This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this
material constitutes acceptance of that license and the conditions of use of materials on this site.
Copyright 2006, The Johns Hopkins University and Karl Broman. All rights reserved. Use of these materials
permitted only in accordance with license rights granted. Materials provided “AS IS”; no representations or
warranties provided. User assumes all responsibility for use, and all liability related thereto, and must independently
review all materials for accuracy and efficacy. May contain materials owned by others. User is responsible for
obtaining permissions for use from third parties as needed.
Estimation
Goal: Estimate a population parameter, θ
Data: X1, X2, . . . , Xn ∼ iid with distribution depending on θ
If one has many estimators to choose from, pick
• That with the smallest SE, among all unbiased estimators
• That with the smallest RMS error, even if biased
Sometimes it’s not clear how to form even one good estimator.
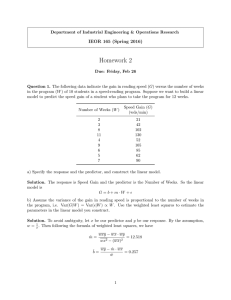
Example 1
Consider the problem of estimating the recombination fraction
(call it θ) between two genetic markers in an intercross.
a a
b b
A A
B B
a a
b b
A a
B b
A a
B b
a A a A a A a A A a A A a a
b B b B b B b B B b B B b b
Note: We won’t observe the haplotypes.
a A
b B
a A
b B
Example 1
Data
AA
BB 58
Bb 8
bb 1
Probabilities
Aa
9
95
12
aa
0
14
53
AA
BB 14 (1 − θ)2
Bb 12 θ(1 − θ)
1 2
bb
θ
4
1
2
Aa
1
2 θ(1 − θ)
[θ2 + (1 − θ)2]
1
θ(1 − θ)
2
aa
1 2
4 θ
1
θ(1 − θ)
2
1
(1 − θ)2
4
Question: Possible estimates of the recombination fraction, θ?
Maximum likelihood estimation
Likelihood function:
Log likelihood:
L(θ) = Pr(data | θ)
l(θ) = log Pr(data | θ)
Maximum likelihood estimate:
Choose, as the estimate of θ, the value of θ
for which L(θ) is maximized.
For the example,
1
1
(9+8+14+12)
2 (58+53)
L(θ) ∝ 4 (1 − θ)
×
×
θ(1 − θ)
1 2(1+0)
1 22
95
× 2 [θ + (1 − θ)2]
4 θ
Example 1: Log likelihood function
−400
log likelihood
−450
MLE = 9.4%
−500
−550
0.0
0.1
0.2
0.3
0.4
0.5
Recombination Fraction
A closer view
−402
log likelihood
−404
MLE = 9.4%
−406
−408
0.06
0.08
0.10
Recombination Fraction
0.12
0.14
A comparison of two estimators
A simple estimator:
Simple estimator
Assume double-heterozygotes are
non-recombinant
For the case n = 250, θ = 0.10;
the results of 1000 simulations:
0.06
0.08
0.10
0.12
0.14
0.12
0.14
Estimate
Simple
Bias –0.005
SE
0.013
RMSE 0.014
MLE
0.000
0.014
0.014
MLE
0.06
0.08
0.10
Estimate
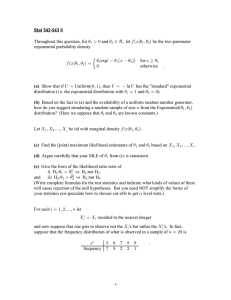
Example 2
Suppose x ∼ binomial(n, p).
log likelihood function: l(p) = log
n
x
px (1 − p)(n−x)
= x log(p) +(n − x) log(1 − p)+ constant
p̂ = x/n
MLE: the obvious thing:
log likelihood
n=100, x=22
−4
MLE = 0.22
−6
−8
0.10
0.15
0.20
0.25
p
0.30
0.35
0.40
Example 3
Suppose x1, . . . , x20 ∼ iid Poisson(λ).
−λ xi
e
λ
/
x
!
i
i
P
= . . . = −20λ + ( xi) logλ + constant
log likelihood function: l(λ) = log
MLE: the obvious thing:
Q
λ̂ = x̄
log likelihood
n=20, mean=1.5
−5
−10
MLE = 1.5
−15
−20
0.5
1.0
1.5
2.0
2.5
3.0
3.5
4.0
λ
Example 4
Suppose x1, . . . , xn ∼ iid N(µ, σ )
log likelihood function: l(µ, σ ) = log
nQ
MLEs: almost the obvious things:
pP
µ̂ = x̄
σ̂ =
(xi − x̄)2/n
√1
i σ 2π
exp
h
− 12
x−µ 2
σ
io
Example 4: the log likelihood surface
n=100
7
6
σ
5
MLE
4
3
9
10
11
12
µ
About MLEs
Maximum likelihood estimation is a general procedure for finding
a reasonable estimator
• In many cases, the MLE turns out to be the obvious thing.
• MLEs are often good (but not necessarily the best) estimators
– Nearly unbiased
– small SE
• Sometimes obtaining the MLEs requires hefty computation
Example 5: ABO blood groups
Phenotype
Genotype
Frequency
O
OO
p2O
A
AA or AO
p2A + 2pApO
B
BB or BO
p2B + 2pB pO
AB
AB
2pApB
Frequencies under the assumption of Hardy-Weinberg equilibrium.
Example 5: Data
Phenotype
No. subjects
% subjects
O
117
46.8%
A
98
39.2%
B
29
11.6%
AB
6
2.4%
Total
250
100%
Question: Estimates of pA, pB , pO ?
Example 5: Estimates
Simple estimates
√
p̃O = 0.468 = 0.684
p̃A to solve p̃2A + 2p̃A0.684 = 0.392
=⇒ p̃A = 0.243
p̃B = 0.024/(2p̃A) = 0.072
Log likelihood:
l(pO , pA, pB ) =
117 log(p2O )+ 98 log(p2A +2pApO )+ 29 log(p2B +2pB pO )+ 6 log(2pApB )
Example 5: log likelihood
1.0
p0 = 0.690
0.8
pA = 0.237
pB = 0.073
pA
0.6
0.4
MLE
0.2
0.0
0.0
0.2
0.4
0.6
pO
0.8
1.0