cytochrome b Alouatta guariba

ABSTRACT
We analyzed mtDNA sequences of the cytochrome b gene from brown howler monkeys living in the Atlantic
Coastal forest in Brazil. Our samples derive from populations from three different localities: Rio de Janeiro state in the north, Santa Catarina state in the south and São Paulo state that lies in an intermediate geographic position. We used maximum parsimony and neighbor joining methods to analyze the cytochrome b genetic variation to make inferences about the intraspecific genealogical relationships of howler monkeys from these three populations. Our findings indicate that cytochrome b gene variation is structured into two main clades. Among the three populations, the cytochrome b sequences from
Santa Catarina and Rio de Janeiro each form separate clades, with no shared diversity. However, the sequences from monkeys from São
Paulo are represented in both of these clades. We hypothesize that the
Santa Catarina and Rio de Janeiro populations underwent a forced separation related to fragmentation of the Atlantic Forest perhaps during the
Pleistocene Epoch as has been hypothesized for other species. We also hypothesize that the São Paulo population formed at a subsequent time following environmental change and forest restoration, thereby accounting for its shared variation with both northern and southern populations.
ACKNOWLEDGEMENTS
We thank Dr. Zelinda Braga,
Projeto Bugio
(http://www.furb.br/especiais/interna.php?se
cao=878) & the QCC Chemistry department for printing our poster.
Poster Design & Printing by Genigraphics ® - 800.790.4001
mtDNA sequences ( cytochrome b ) point to north/south subdivision between populations of brown howler monkeys ( Alouatta guariba ) in the Atlantic Coastal Forest of Brazil.
INTRODUCTION
The brown howler monkey ( Alouatta guariba ) is a medium sized, leaf eating, tree dwelling monkey that resides in Atlantic Coastal Forest, with a wide distribution ranging from southern Bahia, through the coastal Brazilian states, and into the northernmost provinces of Argentina (see figures 1 and 2; Kinzey
1982). We have made inferences regarding intraspecific genealogical relationships of brown howler monkeys by analyzing differences in the mtDNA encoded cytochrome b gene ( cyt b ) in individuals from different locations within the species geographic distribution. This is the first step in assessing the level of population differentiation within the species. The cyt b gene was selected because of its relatively high mutation rate allowing us to analyze individual as well as population level differences in the cyt b gene. We acquired sequences from three different regions of the Atlantic Coastal Forest including the northern state of Rio de Janeiro, in the southern state of Santa Catarina, and also in the state of São Paulo, which lies in an intermediate geographic position between the other two states.
MATERIALS AND METHODS
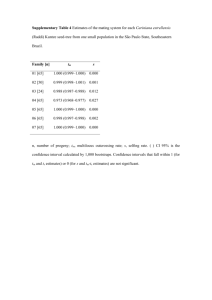
All 19 cyt b gene sequences analyzed in this study were collected in Brazil for an earlier study (Harris et al, 2005). The individuals from which these sequences derive all come from the Eastern Atlantic
Coast of Brazil. They are categorized as the following: five come from the state of Rio De Janeiro, six from the state of São Paulo, and the remaining eight individuals are from the state of Santa Catarina
(see map, fig. 2). We used sequences from both forward and reverse strands to verify ambiguous
DNA bases. We aligned the cyt b sequences by eye using the BioEdit v.7.0.5.3 (Hall, 2005) software. For genealogical analyses, we used the software package MEGA v.4.0.2 (Kumar, 2008) to identify phlyogenetically informative sites. We then employed both maximum parsimony (MP) and neighbor joining
(NJ) methods to build and/or evaluate genealogical trees for the cyt b sequences.
Valerie J. Renard and Eugene E. Harris, Department of Biological Sciences and Geology,
Queensborough Community College, City University of New York
222-05 56
th
avenue
Bayside, NY 11364
RESULTS
Here we present the tree built using the Neighbor Joining
(NJ) method along with bootstrap support for each node described (see fig 3). The tree obtained using the maximum parsimony (MP) method is essentially the same indicating confidence in the cyt b gene tree. The tree reveals the separation of cyt b variation into two main clades, with relatively high bootstrap support for these clades. Among the three populations, the cyt b sequences from Santa Catarina and Rio de Janeiro each form two separate clades (marked on the tree by brackets), with no shared diversity between them. However, the sequences from brown howler monkeys from São Paulo are represented in both of these clades.
Figure 1.
Brown howler monkey (or “Bugio,” as local
Brazilians call it) in its natural forest habitat.
DISCUSSION AND CONCLUSIONS
Figure 1.
Brown howler monkey being treated for broken leg.
Figure 2.
Map representing Atlantic coastal
Brazilian states. Markers are represented
Historically, brown howler monkeys (
A. guariba
) were likely distributed throughout the Atlantic Forest in Brazil from north to south. The populations of these monkeys were likely to be continuous and interbreeding. However, the distinct clades found in this study for brown howlers from
Rio de Janeiro in the north and Santa Catarina in the south, with no overlapping genetic variation, indicates that the continuous population of brown howlers probably underwent subdivision at some time in the evolutionary past. During the course of this division, little or no gene flow between the northern and southern populations occurred. We hypothesize that the division occurred
RJ=Rio de Janeiro, SP= São Paulo,
SC=Santa Catarina
Figure 3 . Phylogenetic tree using Neighbor Joining method with bootstrap support of clades indicated.
and southern regions and forest living brown howlers were restricted to these regions. At a later time, when the forest expanded again – due presumably to increasingly moist conditions – the populations of brown howlers came back into contact. This renewed contact could account for the overlap of genetic diversity from the north and south observed in populations of brown howlers from São Paulo, which lies in an geographically intermediate position.
It is important to note that our study analyzed a single gene
(
cyt b
). Thus, in the future it will be necessary to analyze additional genes to further test our hypothesis of population subdivision. Nevertheless, results from genetic studies of frogs, snakes and birds from the Atlantic Forest show very similar patterns of north/south subdivision, lending support to our historical explanation.