Visualizing Clinical Trial Design Trends by Mapping the
advertisement

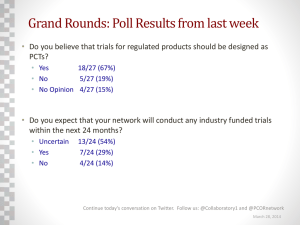
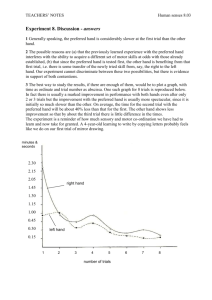
Visualizing Clinical Trial Design Trends by Mapping the Alzheimer’s Disease Trial Space Timothy Schultz M.S., Chaomei Chen Ph.D., Neal Handly M.D. BACKGROUND METHODOLOGY (cont.) CURRENT PROGRESS (cont.) • Ultimate goal is optimization of clinical trial design and evaluation in light of rising cost and failure rate [1] • Lack of historical understanding of therapeutic space, what has been done, what has worked, and what has failed • Research aims to provide a novel approach to identify promising clinical trial design patterns across time by qualifying and quantifying the evolution of clinical trials within a therapeutic space • Utilize a graph-based nonnegative Matrix Factorization (NMF) approach for identifying latent themes based on annotated graph [3] • Calculate trial similarities based on projected lower-dimensional space • Sort similarities temporally to establish direction of influence between similar clinical trials, resulting in a weighted directed graph • Calculate graph metrics over sub-graphs obtained by expanding time window (yearly), such as betweenness centrality and PageRank • Utilize Kleinberg’s Burst Detection algorithm (Fig.2) against graph metrics within each window to identify emergence of pivotal clinical trials • Utilizing numerous visual tools to qualify dynamics of therapeutic area: • Sankey diagrams to visualize change in graph dynamics over time windows [4] • Swim-lane timeline visualizations to visualize emergence of pivotal clinical trials • Interactive graphs for exploration of conceptual graph space • Ability to export to other applications, such as CiteSpace [5] (Fig. 4) PURPOSE AND RATIONALE • Past work has focused on clustering similar clinical trials [2] • Latent topical structures are contained within plain-text documents such as clinical trial protocols, which are pervasive throughout time • Plain-text protocol repositories (e.g. ClinicalTrials.gov) can be extended from a simple document repository by providing analytical capabilities and insights • Enhance ability to recognize and exploit dependency of key clinical trial design variables based on past experience • Ultimate goal is to provide insights into the temporal dynamics of research being conducted within therapeutic spaces METHODOLOGY • Semantically annotate protocol corpus with biomedical ontologies • Express each document as a weighted graph of concepts (Fig. 1) CURRENT PROGRESS 0 0 0 0 Semantically annotated documents Figure 2. Kleinberg’s Burst Algorithm reveals influential drivers of change within the network across periods of time. Express documents as a weighted graph of concepts (affinity matrix) Vector of cooccurring concepts (edge list) • Utilizing Titan graph storage to capture discovered connections between clinical trials and biomedical concepts • Developed web application (D3, Sigma JS) to provide front-end tools to explore therapeutic area (Fig. 3) NCT000 , NCT001, 0.85 NCT002 , NCT003, 0.75 … NCT00X , NCT00Y, 0.98 Calculate similarities based on lower dimensional space, temporally sort Establish graph of interconnected clinical trials 2003 2004 2005 … 2014 Calculate sub-graph dynamics over expanding time windows Figure 1. A graphical overview of the process for identifying thematic clusters of thought within a therapeutic space. drexel.edu/cci FUTURE DIRECTION • Anticipate ability to map underlying ontological concepts to biomedical endpoints found within publicly-available observational datasets (i.e. Alzheimer’s Disease Neuroimaging Initiative) • Quantify within and between cluster variance of endpoints to further visualize influence key clinical trial design considerations have on prospective patient populations over time REFERENCES Dimensionality reduction via NMF Utilize a suite of tools to visualize the dynamics of a therapeutic area (i.e. Sankey Diagrams) Figure 4. Identifying key themes at varying levels of granularity. Shift in treatment from targeting moderate AD (early 2000’s) to more recently early AD (2012) is apparent. Figure 3. Querying and exploring the connections which exist between similar clinical trial protocols. 1. S. L. Mercer, B. J. DeVinney, L. J. Fine, L. W. Green, and D. Dougherty, “Study Designs for Effectiveness and Translation Research: Identifying Trade-offs,” Am. J. Prev. Med., vol. 33, no. 2, pp. 139–154.e2, Aug. 2007. 2. M. R. Boland, R. Miotto, J. Gao, and C. Weng, “Feasibility of feature-based indexing,clustering, and search of clinical trials: A case study of breast cancer trials from ClinicalTrials.gov,” Methods Inf. Med., vol. 52, no. 5, pp. 382–394, Oct. 2013. 3. C.-J. Lin, “Projected Gradient Methods for Nonnegative Matrix Factorization,” Neural Comput, vol. 19, no. 10, pp. 2756–2779, Oct. 2007. 4. M. Rosvall and C. T. Bergstrom, “Mapping Change in Large Networks,” PLoS ONE, vol. 5, no. 1, p. e8694, Jan. 2010. 5. M. B. Synnestvedt, C. Chen, and J. H. Holmes, “CiteSpace II: Visualization and Knowledge Discovery in Bibliographic Databases,” AMIA. Annu. Symp. Proc., vol. 2005, pp. 724–728, 2005. drexel.edu/cci