Lab 11 Key
advertisement

Lab 11 Key
a)
Fit an ordinary MLR model to these data via ordinary least squares. Also fit Lasso,
Ridge and Elastic Net (with say $ $=.5) linear models to these data (i.e. use penalized
least squares and "shrink" the predictions toward ybar and the size of the regression
coefficients). Use glmnet and cross-validation to choose the values of$ $ you employ.
#Here is Code for Lab #11
#Load the psych package
library(psych)
Airfoil<-read.table(file.choose(),header=FALSE,sep='\t')
Air<-data.frame(Airfoil)
names(Air)<-c("Freq","Angle","Chord","Velocity","Displace","Pressure")
summary(Air)
x=as.matrix(Air[,1:5])
y=as.matrix(Air[,6])
#Load the glmnet,stats, and graphics packages and do some
#cross-validations to pick best lambda values
library(glmnet)
## Loading required package: Matrix
## Loading required package: foreach
## Loaded glmnet 2.0-2
library(stats)
library(graphics)
AirLasso<-cv.glmnet(x,y,alpha=1)
AirRidge<-cv.glmnet(x,y,alpha=0)
AirENet<-cv.glmnet(x,y,alpha=.5)
plot(AirLasso)
plot(AirRidge)
plot(AirENet)
#The next commands give lambdas usually recommended .... they are not
#exact minimizers of CVSSE, but ones "close to" the minimizers
#and associated with somewhat less flexible predictors
AirLasso$lambda.1se
## [1] 0.288921
AirRidge$lambda.1se
## [1] 1.193825
AirENet$lambda.1se
## [1] 0.5265081
b)
How do the fits in a) compare? Do residual plots make you think that a predictor linear
in the 5 input variables will be effective? Explain. Does it look like some aspects of the
relationship between inputs and the response are unaccounted for in the fitting? Why
or why not?
Due to figures, it is obvious that they are somehow the same. There is not a
considerable difference between plots. Hence, it shows that all methods are trying to
shrink everything over ybar (the baseline). It is favorable for residuals to randomly
go up and down of baseline and y and yhat has a kind of linear correlation. What we
can see in plots there is a line which is in small y’s small yhats are obtained and in big
y’s big yhats. In linear model there is a bit curvature in model which shows that
something is not considered in model. Linear line is more predictable. (Answer from
Zahra Davoudi)
##
##
##
##
##
##
##
##
6 x 1 sparse Matrix of class "dgCMatrix"
1
(Intercept) 131.64263394
Freq
-0.00109955
Angle
-0.26314173
Chord
-27.78417475
Velocity
0.07259511
Displace
-156.79039132
##
##
##
##
##
##
##
##
6 x 1 sparse Matrix of class "dgCMatrix"
1
(Intercept) 1.308044e+02
Freq
-1.005557e-03
Angle
-2.541206e-01
Chord
-2.582854e+01
Velocity
7.535086e-02
Displace
-1.479461e+02
##
##
##
##
##
##
##
##
6 x 1 sparse Matrix of class "dgCMatrix"
1
(Intercept) 1.312905e+02
Freq
-1.053203e-03
Angle
-2.424408e-01
Chord
-2.632966e+01
Velocity
6.983460e-02
Displace
-1.550157e+02
##
(Intercept)
xFreq
xAngle
xChord
## 1.328338e+02 -1.282207e-03 -4.219117e-01 -3.568800e+01
##
xDisplace
## -1.473005e+02
##
##
##
##
##
##
##
c)
(Intercept)
xFreq
xAngle
xChord
xVelocity
xDisplace
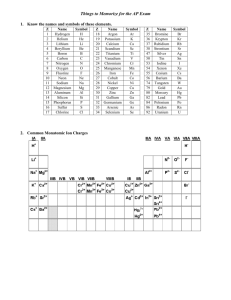
OLS
132.8338
-0.001282207
-0.4219117
-35.688
0.09985404
-147.3005
Lasso
?
?
?
?
?
?
Ridge
?
?
?
?
?
?
xVelocity
9.985404e-02
ENet(.5)
?
?
?
?
?
?
Standardize the input variables and fit k -nearest neighbor predictors to the data for
k=1,3,4,5 (I don't know why the routine crashes when one tries k=2) using knn.reg()
from the FNN package. The " PRESS " it produces is the LOO cross-validation error sum
of squares and can thus be used to choose a value of the complexity parameter k .
Which value seems best? Large k corresponds to which, a "simple" or a "complex"
predictor? How does knn prediction seem to compare to linear prediction in this
"fairly large N and small p " context?
K=3 seems to be the best because it has the least PRESS (LOOCVSSE). K’s shows that
how many nearest vectors you used to predict the value. So, increasing the k makes
the model less complex cause, you have more data to predict with. Also KNN3 is
shinked to ybar (baseline). So, the prediction is easier. Thus, KNN3is selected as the
best model.
#Now do some nearest neighbor regressions
#First, load the FNN package and standardize the predictors
scale.x<-scale(x)
x[1:10]
##
[1]
800 1000 1250 1600 2000 2500 3150 4000 5000 6300
scale.x[1:10,]
##
## [1,]
## [2,]
## [3,]
## [4,]
## [5,]
## [6,]
## [7,]
## [8,]
## [9,]
## [10,]
Freq
-0.6618024
-0.5983622
-0.5190619
-0.4080415
-0.2811610
-0.1225604
0.0836204
0.3532414
0.6704426
1.0828042
Angle
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
-1.146021
Chord
1.798701
1.798701
1.798701
1.798701
1.798701
1.798701
1.798701
1.798701
1.798701
1.798701
Velocity
1.312498
1.312498
1.312498
1.312498
1.312498
1.312498
1.312498
1.312498
1.312498
1.312498
Displace
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
-0.6445901
library(FNN)
Air.knn1<-knn.reg(scale.x,test=NULL,y,k=1)
Air.knn1
## PRESS = 10709.82
## R2-Predict = 0.8501754
Air.knn1$pred[1:10]
##
##
[1] 125.201 126.201 125.201 125.951 127.591 127.461 125.571 121.762
[9] 119.632 118.122
plot(y,y-Air.knn1$pred)
abline(a=0,b=0)
Air.knn3<-knn.reg(scale.x,test=NULL,y,k=3)
Air.knn3
## PRESS = 7573.019
## R2-Predict = 0.8940575
Air.knn3$pred[1:10]
##
##
[1] 126.2477 126.5810 126.3310 126.2043 126.3710 125.7247 124.0080
[8] 122.7547 121.4850 119.6850
plot(y,y-Air.knn3$pred)
abline(a=0,b=0)
Air.knn4<-knn.reg(scale.x,test=NULL,y,k=4)
Air.knn4
## PRESS = 10938.42
## R2-Predict = 0.8469774
Air.knn4$pred[1:10]
##
##
[1] 126.4663 126.4338 126.6135 126.2035 126.4387 126.1912 124.1340
[8] 122.9140 120.9990 118.7973
plot(y,y-Air.knn4$pred)
abline(a=0,b=0)
Air.knn5<-knn.reg(scale.x,test=NULL,y,k=5)
Air.knn5
## PRESS = 11503.44
## R2-Predict = 0.8390731
Air.knn5$pred[1:10]
##
##
[1] 126.3714 126.5714 126.4652 126.2892 126.4774 126.2814 124.7994
[8] 122.2576 120.4236 118.4680
plot(y,y-Air.knn5$pred)
abline(a=0,b=0)
d)
Use the tree package and fit a good regression tree to these data. Employ costcomplexity pruning and cross-validation to pick this tree. How many final nodes do
you suggest? How do the predictions for this tree compare to the ones in a) and c) ?
Better result for this tree according to the residuals. NODE=18.
#Load the tree package
#Do fitting of a big tree
library(tree)
Airtree<-tree(Pressure ~.,Air,
control=tree.control(nobs=1503,mincut=5,minsize=10,mindev=.005))
summary(Airtree)
##
## Regression tree:
## tree(formula = Pressure ~ ., data = Air, control = tree.control(nobs =
1503,
##
mincut = 5, minsize = 10, mindev = 0.005))
## Number of terminal nodes: 32
## Residual mean deviance: 12.42 = 18270 / 1471
## Distribution of residuals:
##
Min.
1st Qu.
Median
Mean
3rd Qu.
Max.
## -11.48000 -2.11700
0.09367
0.00000
2.38700 14.93000
Airtree
## node), split, n, deviance, yval
##
* denotes terminal node
##
##
1) root 1503 71480.00 124.8
##
2) Freq < 3575 1079 34610.00 126.6
##
4) Displace < 0.0155759 769 17080.00 127.9
##
8) Chord < 0.127 331 6638.00 129.7
##
16) Freq < 715 80 2368.00 126.9
##
32) Chord < 0.0762 51
957.50 124.4
##
64) Displace < 0.0135486 39
500.40 122.9
##
65) Displace > 0.0135486 12
86.88 129.2
##
33) Chord > 0.0762 29
483.40 131.5 *
##
17) Freq > 715 251 3496.00 130.5 *
##
9) Chord > 0.127 438 8554.00 126.5
##
18) Freq < 1800 312 4517.00 127.9
##
36) Angle < 3.5 161 1493.00 126.6
##
72) Freq < 565 47
410.50 123.6 *
##
73) Freq > 565 114
477.30 127.9 *
##
37) Angle > 3.5 151 2491.00 129.3
##
74) Velocity < 47.55 80 1066.00 127.7 *
##
75) Velocity > 47.55 71
995.50 131.0 *
##
19) Freq > 1800 126 1847.00 123.0
##
38) Displace < 0.0044289 75
566.30 125.0 *
##
39) Displace > 0.0044289 51
524.80 120.0 *
##
5) Displace > 0.0155759 310 13140.00 123.4
##
10) Displace < 0.0505823 284 10690.00 124.1
##
20) Freq < 1425 196 7077.00 125.7
##
40) Displace < 0.0174419 27 1459.00 120.5
##
80) Freq < 565 15
121.60 114.9 *
##
81) Freq > 565 12
290.40 127.4 *
##
41) Displace > 0.0174419 169 4771.00 126.5
##
82) Velocity < 47.55 81 1855.00 125.0 *
##
83) Velocity > 47.55 88 2546.00 127.9
##
166) Angle < 12.45 36
813.40 130.6
##
332) Freq < 565 20
170.40 133.9 *
##
333) Freq > 565 16
154.10 126.5 *
##
167) Angle > 12.45 52 1286.00 126.1 *
##
21) Freq > 1425 88 2099.00 120.7
##
42) Chord < 0.0762 48
735.90 123.6
##
84) Displace < 0.0286223 36
233.20 125.3
##
85) Displace > 0.0286223 12
65.23 118.3
##
43) Chord > 0.0762 40
491.40 117.2 *
##
11) Displace > 0.0505823 26
585.70 115.3 *
##
3) Freq > 3575 424 25370.00 120.4
##
6) Displace < 0.00156285 112 3477.00 129.4
##
12) Freq < 7150 54
729.90 132.9
##
24) Displace < 0.00107075 36
108.90 135.0 *
##
25) Displace > 0.00107075 18
131.30 128.6 *
##
13) Freq > 7150 58 1509.00 126.2
##
26) Displace < 0.00107075 43
820.60 128.1 *
*
*
*
*
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
27) Displace > 0.00107075 15
120.70 120.9 *
7) Displace > 0.00156285 312 9602.00 117.2
14) Displace < 0.0353012 292 7649.00 117.8
28) Freq < 5650 147 3388.00 120.1
56) Displace < 0.00467589 68
953.50 122.8
112) Chord < 0.1905 32
387.40 125.3 *
113) Chord > 0.1905 36
193.50 120.6 *
57) Displace > 0.00467589 79 1514.00 117.8
114) Chord < 0.0381 16
226.70 123.3 *
115) Chord > 0.0381 63
675.90 116.4 *
29) Freq > 5650 145 2746.00 115.5
58) Displace < 0.00259924 39
461.60 118.6 *
59) Displace > 0.00259924 106 1768.00 114.4
118) Chord < 0.0381 15
213.40 120.5 *
119) Chord > 0.0381 91
898.50 113.4 *
15) Displace > 0.0353012 20
169.00 108.1 *
plot(Airtree)
text(Airtree,pretty=0)
#One can try to find an optimal sub-tree of the tree just grown
#We'll use cross-validation based on cost-complexity pruning
#Each alpha (in the lecture notation) has a favorite number of
#nodes and not all numbers of final nodes are ones optimizing
#the cost-complexity
#The code below finds the alphas at which optimal subtrees change
cv.Airtree<-cv.tree(Airtree,FUN=prune.tree)
names(cv.Airtree)
## [1] "size"
"dev"
"k"
"method"
cv.Airtree
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
$size
[1] 32 31 30 29 26 25 24 23 21 19 18 17 15 14 13 11 10
[24] 1
$dev
[1]
[8]
[15]
[22]
$k
[1]
[7]
[13]
[19]
31132.98
31132.98
34228.70
45289.63
31132.98
31132.98
35228.85
48724.11
-Inf
489.6232
850.3184
1515.5737
8
7
6
4
31132.98 31132.98 31132.98 31132.98 31132.98
31132.98 31132.98 32564.49 34376.07 34454.97
37390.42 39682.32 39682.32 43422.31 45065.73
71589.01
370.2215
567.4349
871.6015
1783.3467
372.5295
568.9482
920.6570
1872.4868
429.3051
586.1602
947.0603
2038.2066
$method
[1] "deviance"
attr(,"class")
[1] "prune"
9
"tree.sequence"
#We can plot SSE versus size of the optimizing trees
plot(cv.Airtree$size,cv.Airtree$dev,type="b")
435.2129
437.4860
611.0563
756.4097
1237.9701 1509.5330
4391.7714 11895.0527
3
#And we can plot versus what the program calls "k," which is the
#reciprocal of alpha from class
plot(cv.Airtree$k,cv.Airtree$dev,type="b")
#Or we can plot versus "1/k"
plot(1/cv.Airtree$k,cv.Airtree$dev,type="b")
#We can see what a pruned tree will look like for a size
#identified by the cross-validation
Airgoodtree<-prune.tree(Airtree,best=15)
Airgoodtree
## node), split, n, deviance, yval
##
* denotes terminal node
##
## 1) root 1503 71480.0 124.8
##
2) Freq < 3575 1079 34610.0 126.6
##
4) Displace < 0.0155759 769 17080.0 127.9
##
8) Chord < 0.127 331 6638.0 129.7 *
##
9) Chord > 0.127 438 8554.0 126.5
##
18) Freq < 1800 312 4517.0 127.9 *
##
19) Freq > 1800 126 1847.0 123.0 *
##
5) Displace > 0.0155759 310 13140.0 123.4
##
10) Displace < 0.0505823 284 10690.0 124.1
##
20) Freq < 1425 196 7077.0 125.7
##
40) Displace < 0.0174419 27 1459.0 120.5
##
80) Freq < 565 15
121.6 114.9 *
##
81) Freq > 565 12
290.4 127.4 *
##
41) Displace > 0.0174419 169 4771.0 126.5 *
##
21) Freq > 1425 88 2099.0 120.7
##
42) Chord < 0.0762 48
735.9 123.6 *
##
43) Chord > 0.0762 40
491.4 117.2 *
##
11) Displace > 0.0505823 26
585.7 115.3 *
##
3) Freq > 3575 424 25370.0 120.4
##
6) Displace < 0.00156285 112 3477.0 129.4
##
12) Freq < 7150 54
729.9 132.9 *
##
13) Freq > 7150 58 1509.0 126.2 *
##
7) Displace > 0.00156285 312 9602.0 117.2
##
14) Displace < 0.0353012 292 7649.0 117.8
##
28) Freq < 5650 147 3388.0 120.1
##
56) Displace < 0.00467589 68
953.5 122.8 *
##
57) Displace > 0.00467589 79 1514.0 117.8 *
##
29) Freq > 5650 145 2746.0 115.5 *
##
15) Displace > 0.0353012 20
169.0 108.1 *
summary(Airgoodtree)
##
##
##
##
##
##
##
##
##
Regression tree:
snip.tree(tree = Airtree, nodes = c(56L, 41L, 42L, 12L, 13L,
18L, 29L, 57L, 19L, 8L))
Variables actually used in tree construction:
[1] "Freq"
"Displace" "Chord"
Number of terminal nodes: 15
Residual mean deviance: 18.56 = 27620 / 1488
Distribution of residuals:
##
Min.
## -13.5800
1st Qu.
-2.7420
Median
0.2656
Mean
0.0000
3rd Qu.
2.7580
plot(Airgoodtree)
text(Airgoodtree,pretty=0)
Airpred<-predict(Airgoodtree,Air,type="vector")
plot(y,y-Airpred)
abline(a=0,b=0)
Max.
14.4800
e) Fit a random forest to these data using m=2 (which is approximately the standard
default of the number of predictors divided by 3) using the randomForest package. How do
these predictions compare to all the others?
These predictions are similar to all the others.
#Next is some code for Random Forest Fitting
#First Load the randomForest package
library(randomForest)
##
##
##
##
##
##
##
##
randomForest 4.6-10
Type rfNews() to see new features/changes/bug fixes.
Attaching package: 'randomForest'
The following object is masked from 'package:psych':
outlier
Air.rf<-randomForest(Pressure~.,data=Air,
type="regression",ntree=500,mtry=2)
Air.rf
##
## Call:
## randomForest(formula = Pressure ~ ., data = Air, type = "regression",
ntree = 500, mtry = 2)
##
Type of random forest: regression
##
Number of trees: 500
## No. of variables tried at each split: 2
##
##
Mean of squared residuals: 4.720337
##
% Var explained: 90.07
predict(Air.rf)[1:10]
##
1
2
3
4
5
6
7
8
## 126.5397 126.5226 126.4388 126.3687 126.2617 125.4725 123.9205 121.8551
##
9
10
## 120.7444 118.3694
f)
g)
People sometimes try to use "ensembles" of predictors in big predictive analytics
problems. This means combining predictors by using a weighted average. See if you
can find positive constants
COLS , CLASSO , CRIDGE , CENET , CKNN , CTREE , CRF
adding to 1 so that the corresponding linear combination of predictions has a larger
correlation with the response variable than any single predictor. (In practice, one
would have to look for these constant inside and not after cross-validations.)
#This code produces a scatterplot matrix for the predictions
#with the "45 degree line" drawn in
comppred<-cbind(y,predict(AirLM,newx=x),predict(AirLasso,newx=x),
predict(AirRidge,newx=x),predict(AirENet,newx=x),
Air.knn3$pred,Air.knn5$pred,Airpred,predict(Air.rf))
colnames(comppred)<-c("y","OLS","Lasso","Ridge",
"ENet(.5)","3NN","5NN","Tree","RF")
pairs(comppred,panel=function(x,y,...){
points(x,y)
abline(0,1)},xlim=c(100,145),
ylim=c(100,145))
round(cor(as.matrix(comppred)),2)
##
##
##
##
##
##
##
##
##
##
y
y
1.00
OLS
0.72
Lasso
0.72
Ridge
0.72
ENet(.5) 0.71
3NN
0.95
5NN
0.92
Tree
0.78
RF
0.96
OLS Lasso Ridge ENet(.5) 3NN 5NN Tree
RF
0.72 0.72 0.72
0.71 0.95 0.92 0.78 0.96
1.00 1.00 1.00
1.00 0.74 0.78 0.73 0.77
1.00 1.00 1.00
1.00 0.73 0.78 0.73 0.77
1.00 1.00 1.00
1.00 0.74 0.78 0.73 0.77
1.00 1.00 1.00
1.00 0.73 0.78 0.73 0.77
0.74 0.73 0.74
0.73 1.00 0.98 0.80 0.97
0.78 0.78 0.78
0.78 0.98 1.00 0.82 0.97
0.73 0.73 0.73
0.73 0.80 0.82 1.00 0.84
0.77 0.77 0.77
0.77 0.97 0.97 0.84 1.00
For question 2, rerun the code with different data set.