Lab 5
advertisement

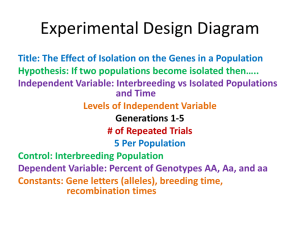
Lab 5 Self Assessment. The data in arabidop.csv are measurements of the alanine concentration in samples from three genotypes of Arabidopsis. This is part of a gene knockout study, but details of that are suppressed. There are four plants from each of three genotypes. It is appropriate to consider plant the experimental unit even though genotype is not randomly assigned to a plant. Each plant was measured twice (technical rep). Before analysis, you need to calculate the average concentration for each plant. 1) Calculate the average concentration in each plant. What is the average concentration in the third WT plant (genotype = WT, rep=3)? 2) Fit a model with a separate mean for each genotype and calculate the residuals for each of the 12 observations. What is the residual for the third WT plant (genotype=WT, rep=3)? Note: In lecture, we’ve only talked about analyzing two groups. The proc glm or lm() or fit Y by X work exactly the same for 3 groups (these data) as they do for 2 groups (what you know about). 3) draw side-by-side box plots of the residuals for the three genotypes. Based on this plot, what could you say about the assumption of equal variances? Answers: 1) 0.3967 (more figures than appropriate so you know whether you’ve done the correct calculations) 2) 0.0636 (again, more figures than appropriate) 3) It seems that the HT genotype is more variable than the other two. 0.4 0.3 0.2 resid 0.1 0 -0.1 -0.2 -0.3 HM HT Genotype WT