Statistics 580 Introduction to Markov Chain Monte Carlo
advertisement

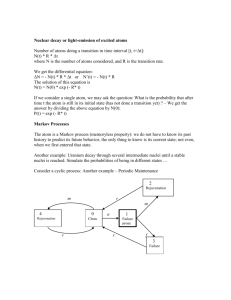
Statistics 580
Introduction to Markov Chain Monte Carlo
Introduction to Markov Chains
A stochastic process is a sequence of random variables {X(t), t ∈ T } indexed by a parameter t in
an index set T. X(t) is called the state of the process at time t and the set of possible realizations of
X(t) defines the state space denoted by S. The time or the parameter space may be discrete, for e.g.,
T = {0, 1, . . .} or continuous, for e.g., T = (0, ∞). The state space may also be discrete, for e.g.,
S = {0, 1, . . .} or continuous, for e.g., S = (−∞, ∞).
A discrete parameter stochastic process {X(t), t = 0, 1, 2. . . .} or a continuous parameter process
{X(t), t ≥ 0} is said to be a Markov process if, for any set of n time points t 1 < t2 < . . . < tn in
the index set T ,
P r[X(tn ) ≤ xn |X(tn−1 ) = xn−1 , . . . , X(t1 ) = x1 ] = P r[X(tn ) ≤ xn |X(tn−1 ) = xn−1 ].
It says that the probability distribution of future states in a Markov process depends only on the present
(or most recently observed) state and not on the past states. Markov processes are classified according
to the nature of the state space or the parameter space. A Markov process whose state space is discrete
is called a Markov chain. We shall use the set {0, 1, . . .} to denote the state space of a Markov chain
and first consider discrete parameter Markov chains where, without loss of generality, we shall use
the notation X0 , X1 , . . . , Xn , . . . to denote the states.
The transition probability function of a discrete parameter Markov chain that define the probability distribution of the next state given the present state is given by,
pij (m, n) = P r(Xn = j|Xm = i)
for all states i and j and n ≥ m. The matrix of transition probabilities is denoted by P (m, n) =
(pij (m, n)). In order to give the probability law of a discrete parameter Markov chain {X n } it is sufficient
to specify, for all times n ≥ m, pj (n) = P (Xn = j) and pij (m, n) for all states i and j. A fundamental
relation satisfied by the transition probability function of a Markov chain is the Chapman-Kolmogorov
equation: for any times n > u > m and states i and j,
pij (m, n) =
X
pik (m, u)pkj (u, n).
k
or, in terms of transition probability matrices,
P (m, n) = P (m, u) P (u, n).
If pij (m, n) depend only on the difference n − m they are said to be stationary transition
probabilities and the Markov chain is said to be stationary or homogeneous. In a stationary Markov
chain, the k-step transition probability function is denoted by
(k)
pij = P r(Xn+k = j|Xn = i)
1
(k)
and the k-step transition probability matrix by P (k) , where P (k) = (pij ). The one-step transition
probabilities of a stationary Markov chain are denoted by
(1)
where by definition
X
pij ≡ pij = P r(Xn = j|Xn−1 = i)
pij = 1 and the matrix of these transition probabilities is the square matrix
j
p11
p
P =
21
..
.
p12
p22
..
.
p13 · · ·
p23 · · · ,
..
.
···
and is called the one-step transition matrix. We see that the rows of P sum to one and hence it
is a stochastic matrix or a marix of probabilities. As an example, in image analysis a binary image is
represented in pixels where black or white pixels are indicated by θi = 1 or 0, respectively. The posterior
joint density of the true image θ given the observed noisy image x (i.e., observed data) is
p(θ|x)
∝
f (x|θ)g(θ)
where f (x|θ) is the model for how the true image is corrupted by noise, and g(θ) is the prior that involves
knowledge about the types of images under consideration. The number of rows (= number of columns)
of P in this example may be 2262,144 > 107,857 .
Examples of discrete parameter Markov chains:
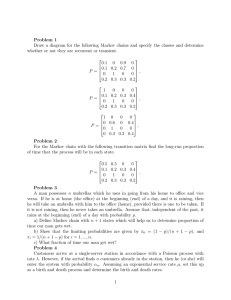
Example 1 - Two-state Markov chains:
A two-state weather model with the two states being “rain” or “no rain”, on successive days. The one-step
transition probability matrix is:
P =
rain
no rain
rain
α
β
no rain!
1−α
1−β
where the elements represent probabilities that it will rain or not on given day conditional on that it rained
or not the previous day. Here the Markov chain will be homogeneous since the transition probabilities
are stationary, since they are unaffected by what day it is.
Example 2:
Toss a coin where P r(head) = p, repeatedly. After the nth toss, let Xn represent the number of heads
that have appeared so far.
Then the one-step transition probability matrix is:
P =
1−p
p
0
...
0
1−p
p
...
0
0
1−p p
..
..
..
..
.
.
.
.
where the elements of the 1st row are
... 0
... 0
... 0
... 0
p11 = P (Xn = 1 | Xn−1 = 1) ,
p12 = P (Xn = 2 | Xn−1 = 1) ,
p13 = P (Xn = 3 | Xn−1 = 1) , ..., etc.
2
and the elements of the 2nd row are
p21
p22
p23
p24
= P (Xn
= P (Xn
= P (Xn
= P (Xn
=1
=2
=3
=4
|
|
|
|
Xn−1
Xn−1
Xn−1
Xn−1
= 2) ,
= 2) ,
= 2) ,
= 2) , ..., etc.
We see that Xn = Xn−1 + S where S ∼ Bernoulli and Xn−1 and S are independent, which implies that
{Xn } is a Markov chain.
Some definitions and results about discrete parameter Markov chains
Recall that the k-step transition probability function was defined as
(k)
pij = P r(Xn+k = j | Xn = i)
for any integer n. The notation
Xn+k = j | Xn = i
says that X goes from state i to state j in k steps in time. P (k) denotes the k-step transition matrix,
(k)
whose elements are pij , and P is the one-step transition matrix whose elements are pij . For a stationary
Markov chain
P (k) = P k
p(k) = p(0) P k
where
p(k)
(k)
p1
(k)
p2
..
.
=
(k)
p
j
..
.
,
p(0)
p1
p2
..
.
(k)
, pj = P r(Xn+k = j),
=
pj
and, pj = P r(Xn = j),
..
.
(1)
(0)
are the k-step unconditional probabilities. Note that pj ≡ pj is the pmf of the random variable Xn , pj
is the pmf of the random variable Xn+1 , etc. These results are immediate from the Chapman-Kolmogorov
equations. Consequently, the probability law of a homogeneous Markov chains is completely determined
once one knows the one-step transition probability matrix P and the unconditional probability vector
p(0) at time 0.
Example:
The two-state weather model with α = 0.7, β = 0.4 is stationary. Thus it follows that
P =
0.7 0.3
0.4 0.6
!
2
P =
0.61 0.39
0.52 0.48
!
4
P =
0.575 0.425
0.567 0.433
!
Note that the rows of P 4 are almost identical showing that the probability of rain or no rain on a certain
day does not depend on whether it rained or not four days earlier.
3
Some properties of Markov chains:
The following definitions, properties and results are useful, in general, and for defining certain useful
classes of Markov chains, in particular:
1. Markov chain is irreducible if every state can be reached from every other state. That is, for all
(k )
i, j there exists some k0 such that pij 0 > 0. We say that all states communicate with each other.
Example: The chain described by P on the left is irreducible and the
right is not:
0.5 0.5
0.5 0.5 0.0
0.5 0.5
P =
P =
0.5 0.3 0.2
0.2 0.3
0.0 0.3 0.7
0.0 0.0
chain described by P on the
0.0
0.0
0.3
0.0
0.0
0.0
0.2
1.0
(k)
(k)
2. Define fij ≡ P r(Xn+k = j for the 1st time |Xn = i). That is, fij is the probabilty that Xn+k is
(k)
for the first time in the j th state given that Xn was in the ith state. Thus fjj probability of first
passage from state j to state j in k-steps. The j th state is said to be persistent (i.e.,not transient) if
∞
X
(k)
fjj = 1.
k=1
That is, having started at state j, the probability that the chain will eventually return to j is one.
Note that in the literature the term recurrent is sometimes used in place of the term persistent to
describe states that satisfy this condition.
(k)
3. The j th state is periodic of period tj if pjj > 0 only when k = ν tj where ν is an integer.
4. Markov chain is aperiodic if no states are periodic.
5. The Markov chain is persistent (not transient) if all states are not transient.
Theorem
Consider an irreducible, aperiodic, and persistent Markov chain whose mean recurrence time is finite,
i.e.,
mjj =
∞
X
k=1
(k)
k fjj < ∞ .
An invariant (or limiting) distribution for the Markov chain is said to exist if there exists a probability
distribution {πj } such that,
(k)
lim pij = πj
k→∞
for all j = 1, 2, . . .. If the invariant distribution {πj } exists then it is the unique solution to the equation
πj =
X
i
that satisfies
P
πj = 1.
4
πi pij .
(1)
Equation (1) follows since
(k)
P k = pij
π1 π2 · · ·
k→∞
.
−→
..
π1 π2
and clearly, P k+1 = P k P . Therefore, as k → ∞,
π0
π0
..
.
=
π0
π0
..
.
..
.
π0
P ,
where π 0 = (π1 , π2 , . . . , ), which implies (1). P is said to satisfy global balance if p ij satisfies (1). π is also
known as the equilibrium distribution or the stationary distribution.
The idea behind Markov chain Monte Carlo is to find an appropriate Markov chain (i.e, P ) whose
invariant distribution {πj } is the distribution from which we wish to draw samples. From a starting
realization X0 , simulate X1 according to the transition matrix P subsequently simulate X2 from X1 and
P and so forth. After a burn in
(k)
pij = P r Xk = j|X0 = i ' πj
and so Xk is a realization from the distribution {πj , j = 1, 2, . . .}. How can we find such a Markov chain?
We need at least one more result.
Reversibility of Markov chains
Let {Xn : −∞ < n < ∞} be a Markov chain with invariant distribution {πj }. Then P r(Xn = j) = πj .
Consider the time-reversed process
Zn = X−n
Now {Zn } is a Markov chain with transition probabilities
qij = P r(Zn = j|Zn−1 = i)
= P r(Zn = j , Zn−1 = i)/P r(Xn−1 = i)
= P r(X−n+1 = i|X−n = j)
= pji
P r(X−n = j)
P (X−n+1 = i)
πj
πi
The Markov chain is time reversible if {Zn }
d
=
{Xn } which implies qij = pij which in turn implies
πj pji = πi pij
for all i, j ∈ S.
5
(2)
Proposition
Equation (2) implies Equation (1).
Proof:
R.H.S. of (1)
=
X
πi pij
X
πj pji
i
=
i
= πj = L.H.S. of (1) .
Condition (2) is obviously the stronger condition and the π are said to satisfy detailed balance. To simulate
samples from a distribution {πj }, it is sufficient that a Markov chain is defined via transition probabilities
{pij } that satisfy the relation πi pij = πj pji . This relation is called the reversibility condition. Markov
Chain Monte Carlo turns the theory around: the invariant density is known (perhaps up to a constant
multiple) – actually it is the target density from which samples are desired – but the transition matrix is
unknown. To generate samples from π(.), Markov Chain Monte Carlo methods use a transition matrix
called a nominating matrix and employs an acceptance-rejection algorithm whose nth iterate converges
to π(.) for large n.
Metropolis algorithm (Metropolis, et al., 1953)
Suppose the nominating matrix Q is any symmetric matrix of probabilities i.e., q ij = qji . We would like
to obtain a sample from a distribution {πj } where πj = P r(Xn = j) by generating observations from a
Markov chain that has {πj } as its invariant distribution. Metropolis algorithm starts with proposed state
i and decides whether it moves to a new state j based on Bernoulli trial:
Step
0. Set xn−1 = i where i is any realization from πi .
1. Generate j from the probability distribution {qij ; j = 1, 2, . . .}
2. Set r = πj /πi .
3. If r ≥ 1 set xn = j
Otherwise generate u from U (0, 1)
if u < r set xn = j
else set xn = xn−1
4. Set n = n + 1, go to Step 1 .
In the above algorithm, the value j is accepted with probability αij = min {πj /πi , 1}. The proof that the
equilibrium distribution of the chain constructed by the above algorithm is indeed {π j }, it is sufficient to
check that the detailed balance condition holds.
6
Example: As an application of the Metropolis algorithm, suppose that we want to generate from the
Poisson distribution with mean λ i.e.,
π j = P Xn = j =
1 j −λ
λ e ,
j!
j = 0, 1, . . .
We will use the nominating probability matrix
Q=
1/2 1/2 0
0
1/2 0 1/2 0
0
0 1/2 0 1/2 0
0
0 1/2 0 1/2
..
..
..
..
..
.
.
.
.
.
···
···
···
···
q00 = 1/2
qij = 1/2 for j = i − 1
i.e.,
= 1/2 for j = i + 1
= 0, otherwise
which is symmetric (and is a one-step transition matrix).
The Metropolis algorithm for generating samples from Poisson(6) is as follows:
Step 0. Start with xn−1 = i
Step 1. Generate j from {qij }
if i 6= 0
if i = 0
i.e. , generate u1 from U (0, 1)
(
if u1 < 1/2, set j = i − 1
else set j = i + 1
if u1 < 1/2, set j = 0
else set j = 1
Step 2. Set r = πj /πi = (i! λj )(j! λi )
i.e., set r = 1,
= i/λ,
= λ/j,
if i = 0, j = 0
if j = i − 1
if j = i + 1
Step 3. If r ≥ 1, set xn = j
Otherwise, generate u2 from U (0, 1)
if u2 < r, set xn = j
else, set xn = xn−1
Step 4. Set n = n + 1, go to 1
7
The table below display values of relevant quantities computed in the first 15 iterations of this algorithm for Poisson with λ = 6 starting with x0 = 2:
-------------------------------------------------n
i
u1
j
r
u2
--------------------------------------------------1
2 0.71889082
3 2.0000000 0.83568994
2
3 0.92144722
4 1.5000000 0.67244221
3
4 0.48347869
3 0.6666667 0.23677552
4
3 0.38764000
2 0.5000000 0.70580029
5
3 0.66973964
4 1.5000000 0.47446056
6
4 0.51325076
5 1.2000000 0.44375696
7
5 0.22118260
4 0.8333333 0.79923561
8
4 0.32724500
3 0.6666667 0.55147710
9
3 0.32624403
2 0.5000000 0.88511680
10
3 0.32752058
2 0.5000000 0.82785282
11
3 0.51644296
4 1.5000000 0.30783601
12
4 0.53919790
5 1.2000000 0.40234452
13
5 0.95002276
6 1.0000000 0.07881027
14
6 0.07521049
5 1.0000000 0.25551719
15
5 0.78899123
6 1.0000000 0.59512748
--------------------------------------------------The R code that was used to generate these iterates is as follows:
poisson.metro=function(lamda,i,n)
{
y=seq(n)
for(k in 1:n)
{
u1=runif(1)
j =if(u1<.5)
ifelse(i==0,i,i-1) else i+1
r =switch(i+2-j,lamda/j,1,i/lamda)
u2 =runif(1)
new=if(r>=1)j else
{if(u2<r)j else i}
i=new
y[k]=i
}
return(y)
}
8
Rel.Freq.
Rel.Freq.
0.20
0.10
0.0
0.20
0.10
0.0
•
•
2
•
2
•
4
•
6
j
•
8
•
•
10
•
•
•
4
•
•
6
•
j
•
8
•
•
10
•
Iterates 4505-5000 in steps of 5
•
•
•
Iterates 101-200
•
12
•
12
•
•
•
•
2
•
2
•
4
j
6
•
•
•
4
•
•
6
•
j
•
8
•
•
10
•
•
Iterates 4500-5000 in steps of 3
•
•
•
Iterates 901-1000
Metropolis Algorithm for generating Poisson(6) Samples
Rel.Freq.
Rel.Freq.
0.20
0.10
0.0
0.20
0.10
0.0
9
•
12
8
•
•
•
The last page showed plots of relative frequency barcharts constructed from 100 values obtained from this
Metropolis sampler (each sample plotted obtained as labelled on the plots) superimposed by actual probability mass function of the Poisson(6) shown on the plots with connected line segements for comparison
purposes.
A more general form of Metropolis algorithm was given by Hastings(1970) and is usually referred to as
Metropolis-Hastings algorithm. In this case, qij , the nominating probabilities are more general instead of
being symmetrical. The acceptance probability of j in this case is given by
αij = min {πj qji /πi qij , 1}
Metropolis-Hastings Algorithm (discrete state space case)
Step
0. Set xn−1 = i where i is any realization from πi .
1. Generate j from the probability distribution {qij ; j = 1, 2, . . .}
2. Set r = πj qji /πi qij .
3. If r ≥ 1 set xn = j
Otherwise generate u from U (0, 1)
if u < r set xn = j
else set xn = xn−1
4. Set n = n + 1, go to Step 1 .
It is easily shown that the detailed balance condition holds for this algorithm as well thus proving that
its equilibrium distribution is {πj }.
In both Metropolis and Metrpolis-Hastings the resulting chain would have transition probability matrices
defined by
pij = qij αij , fori 6= j
X
6= iqij αij
pii = 1 −
j
The theory on the discrete parameter Markov chains carry over to the continuous time, continuous state
space case with some theoretical generalizations. In particaular, the transition matrix P becomes a
transition kernel p(x, y) for x, y ∈ <, which can be used to compute probabilities as usual:
P (y ∈ A|X = x) =
Z
A
p(x, y)dy
Other properties need to be defined accordingly, for e.g. recurrence is defined in terms of sets with
positive probability of being visited infinitely often. The stationary or the equilibrium distribution π(y)
of a continuous Markov chain then satisfies
π(y) =
Z
p(x, y) π(x) dx
10
Now we generalize Metropolis-Hastings algorithm to the case when the state space is continuous instead
of discrete. In this case let π(x) denote the invariant distribution of a Markov chain and is the target
density from which samples are desired. Let q(x, y) denote the candidate-generating density, or the
proposal density meaning that when the process is at the point x, a value y is generated from this density.
The Metropolis-Hastings algorithm is described in terms of the acceptance probability (or probability of
a move) α(x, y):
)
(
π(y) q(y, x)
, 1 , if π(x) q(x, y) > 0
α(x, y) = min
π(x) q(x, y)
= 1, otherwise
The idea is that at a current state X(t) = x, a candidate value for the next state y is generated from
q(x, y); this value is accepted as the next state with probability α(x, y). Transition probabilities for the
chain are then given by the density
p(x, y) = q(x, y) α(x, y)
= 1−
Z
q(x, t) α(x, t) dt
if y 6= x
if y = x
The reversibility condition is then
π(x) p(x, y) = π(y) p(y, x)
and if it is satisfied and p(x, y) leads to a irreducible, aperiodic chain, then π(.) will be the invariant
distribution. These conditions are usually satisfied if q(x, y) is positive on the same support as that of
π(·)
Metropolis-Hastings Algorithm
Step 0. Set n = 0 and start with xn
Step 1. Generate y from q(xn , .) and u from U (0, 1)
Step 2. If u ≤ α(xn , y)
Set xn+1 = y
Else
Set xn+1 = xn
Step 3. Set n = n + 1, go to Step 1
n
Step 4. Return x0 , x1 , . . . , xN
o
11
Example 1: Implement a Metropolis-Hastings algorithm to simulate from the mixture
.7N (7, 0.52 ) + .3N (10, 0.52 )
using N (x, 0.12 ) as the proposal distribution. For starting values x0 = 0, 7, and 15 run the chain for
10, 000 iterations. Plot the sample path of the output for each chain. Change the proposal distribution
to improve the convergence properties of the chain.
It is clear that the target π(x) is the density of the mixture of the two normals above. The proposal
density q(x, y is the density of N (x, 0.12 ) given by
1
1 y−x 2
√
)
exp − (
2 .1
2π(.1)
which is symmetric in x and y implying q(x, y) = q(y, x). Thus the acceptance probability is given by
α(x, y) = min
n π(y)
o
,1 .
π(x)
The following R code was used to generate two paths of the chain for starting values 0 = 0.0 and x0 = 7.0,
repectively, the graph of which are on the next page.
normal.metro=function(x0,n)
{
set.seed(1234,"Mersenne-Twister")
r=rep(0,n)
x=x0
for(k in 1:n)
{
u=runif(1)
y=rnorm(1,x,.1)
if(u<alpha(x,y))
{
x=y
}
else
{
x=x0
}
r[k]=x
}
return(r)
}
alpha = function(x,y) {
# Acceptance probability calculation
return( min( 1, (.7*dnorm(y,7,.5)+.3*dnorm(y,10,.5))/(.7*dnorm(x,7,.5)+.3*dnorm(x,10,.5))))
}
12
Proposal sigma=.4; Starting value x=7
9
6.0
6
6.5
7
8
r(t)
7.0
r(t)
7.5
10
8.0
Proposal sigma=.1; Starting value x=7
0
2000
4000
6000
8000
10000
0
2000
4000
t
6000
8000
10000
t
Figure 1: Paths of Random Samples from the Normal Mixture MCMC, respectively
Example 2: (Chib and Greenberg, The American Statistician, (1995))
To illustrate the Metropolis algorithm we consider sampling from the bivariate normal distribution N 2 (µ, Σ),
where
!
1
1 .9
µ=
.
and Σ =
.9 1
2
Note that random variates from the multivariate normal distribution are usually obtained using the Cholesky
factorization Σ = T0 T where T is a unique upper triangular matrix. Generallly, a random vector z is generated
from Np (0, I) and is transformed to Np (µ, Σ) using y = µ + T0 z.
For applying the Metropolis algorithm for this problem, we will re-state the problem as follows: Suppose that
we want to generate from x ∼ N2 (µ, Σ) i.e., π(x) will be the density
1
1
π(x) =
exp − (x − µ)0 Σ−1 (x − µ) ,
1/2
2
2π|Σ|
x ∈ <2 .
Choose the candidate generating
density to be the
pdf of y ∼ N2 (x, D) where D = 0.6 .40 . Notice that
1
q(x, y) = 2π|D|
− 21 (y − x)0 D−1 (y − x) is symmetric in x and y, so that the acceptance probability is
1/2 exp
given by
α(x, y) = min
n exp[− 1 (y − µ)0 Σ−1 (y − µ)]
2
exp[− 12 (x
−1
− µ)0 Σ
13
(x − µ)]
o
,1 ,
x, y ∈ <2
Thus the Metropolis algorithm for generating from π(x) can be described as follows:
Step 0.
Set n = 0 and start with xn , say (1.2, 1.8)T
Step 1. Generate y from q(xn , ·) and u from U (0, 1).
Step 2. If u ≤ α(xn , y)
Set xn+1 = y
Else
Set xn+1 = xn
Step 3. Set n = n + 1, go to step 1
Step 4. Return {x0 , x1 , x2 , . . . , }
As an exercise, we shall implement R functions to generate from π(·) using both the standard algorithm and the
Metropolis algorithm given above and obtain scatterplots as shown on p.334 of Chib and Greenberg.
Usually, the choice of a proposal density (candidate generating density) is problem specific. However, the
simplest choice for q(x, y), in general, is a random walk. That is, given x, y is generated simply using y = x + z,
where z is independently ∼ U (−k, k) where k is a small value, say k = .1, chosen depending on the starting
value. If the random walk is the choice, q(x, y) is symmetric, so the Metroplois Algorithm can be used (instead
of the Metropolis-Hastings version).
14
Monte Carlo Sampling from a Posterior Distribution using Metropolis-Hastings Algorithm
Recall that in Bayesian applications, we would like to sample from the posterior p(θ|y) where:
p(θ|y) ∝ f (y|θ) π(θ)
i.e., “posterior”
is proportional to
“data model”
× “prior”
This allows us to study the posterior distribution or just estimate the posterior mean E(θ|y) empirically and thus
avoid the computation of a complicated integral. The data model is usually the joint density of the observations
(i.e., the likelihood function). Note that if π(·) is a conjugate prior then p(θ|y) can be obtained in closed form.
To sample from p(θ|y) we will restate the Metropolis-Hastings algorithm in the following form:
Let the acceptance probability of moving from θn to θ ∗ be α(θn , θ∗ ) where
∗
α(θn , θ ) = min
(
p(θ∗ |y) q(θ ∗ , θn )
,1
p(θn |y) q(θn , θ∗ )
)
where p(θ|y) is the posterior and corresponds to π(·) in the original description of the M-H algorithm and q(θ n , θ)
corresponds to the candidate-generating density.
Metropolis-Hastings algorithm for sampling from a posterior
Initialize n = 0 and θn
Repeat {
Sample θ ∗ from q(θn , θ)
Sample u from U (0, 1)
If u ≤ α(θn , θ∗ ) then
set θn+1 = θ∗
Else
set θn+1 = θn
Set n = n + 1
}
The implementation of this algorithm for a real problem of sampling from a posterior distributiom of a parameter
is discussed. The data set used consists of measurements of the weight of a block of metal called NB10 used
as a standard for 10g., made by the National Bureau of Standards (now NIST) annually to a high degree of
accuracy. See pages attached at the end of this note. The problem is to estimate the variance of these data using
a Gaussian model, i.e., yi |µ, σ 2 ∼ N (µ, σ 2 ) and a prior (µ, σ 2 ) ∼ h(µ, σ 2 ). First, some implementation concerns.
15
Practical Issues
1. Choosing Initial Values
Metropolis-Hastings requires you to pick just a single initial value θ0 , in many cases this one value may
suffice. It is recommended that you select a value near the center of the posterior from which you are trying
to simulate. This will increase the possibility of the Markov Chain reaching the invariant distribution
reasonably quickly. This value could be obtained from any information you have of the posterior, such as
a good estimate of θ like the maximum likelihood estimate. From a practical viewpoint, a problem with
just starting with a single value is that we will not know in advance whether the chain will be mixing well,
i.e., it is reaching all areas of probability of the posterior distribution. For example, if the posterior is
multimodal the starting near one of the modes the chain may not find the other modes. One strategy to
overcome this is to use several different initial values.
2. Choosing a Convergence Monitoring Strategy
We have two issues to deal with:
• how to decide if the chain has reached equilibrium.
• how to monitor the output from that point onwards to obtain the posterior summaries.
If you started from quite a bit away from the true posterior then the output will be similar to the one
shown in the time series plot of the Gaussian model for the NB10 data. It clear from that graphic that
the chain is not mixing well: there are long periods where it does not move at all. This is caused by the
large first order auto-correlations.
A solution for this problem is to allow a burn-in period: nB , i.e., discard, say the first 1000 (or 5000)
values output and then start observing the time series plot. After burn-in, monitor the output for a larger
number of iterations, upto , say 25,000 to 100,000 iterations. One could also use thinning i.e., retaining
only every 100th or 200th value of the chain thus reducing the auto-correlation to virtually zero.
From this part one could estimate posterior means and standard errors, obtain plots of histograms and
density traces of the marginal posteriors or estimate posterior covariance matrixix. etc.
3. Choosing a Candidate Generating Density(CGD)
This is very difficult problem since Metropolis-Hastings will work for many choices of CGD’s. However,
one may want to select q(x, y) that results in a chain that mixes well.
One strategy is to pick a CGD such that, on the average, a move to the left or the right is equally likely.
That is E(θ ∗ |θt ) = θt where θ ∗ represent a new move and θt is the current value. The use of this strategy
is illustrated below for sampling from the posterior variance of the Gaussian model for the NB10 data.
16
For the NB10 data, pretend that µ for the data distribution is known (assume that it is equal to the sample
mean 404.59), so that the problem reduces to one of studying the posterior distribution of a single parameter,
σ2.
σ 2 ∼ SI −χ2 (νp , σp2 )
Prior:
i.i.d
Data Model: yi |σ 2 ∼ N (µ, σ 2 ),
i = 1, . . . , n
The problem then is to sample from the posterior p(σ 2 |y) using MC.
Note 1: For this problem, we shall ignore the fact that the exact posterior distribution can be derived theoretically:
!
νp σp2 + n s2∗
2
2
σ |y ∼ SI −χ νp + n,
νp + n
where s2∗ =
1
n
n
X
i=1
(yi − µ)2 .
Note 2: We use the short-hand notation SI −χ2 (νp , σp2 ) for the “Scaled Inverse-χ2 ” distribution that is often
used as the conjugate prior for the variance parameter.
To use the M-H algorithm to get MC samples from the posterior distribution p(σ 2 |y) we need to consider the
following implementation details for writing the needed R functions.
Implementation Details of Sampling from the Posterior Distribution of the Variance for NB10
Data
1. Selection
of an appropriate candidate generating density (CGD). Since we know that the prior is SI −
2
2
χ νp , σp , we might consider the CGD q(σn2 , σ 2 ) to be the density of SI−χ2 (ν1 , σ12 ) for some ν1 , σ12 where
ν1 is the degrees of freedom parameter and σ12 is the scale parameter. Note that the density function for
this distribution is
!
!
ν1 /2 −(ν1 /2+1)
ν1 σ12
2
2
2
2
exp −
σ
π σ |ν1 , σ1 = c σ1
2σ 2
with mean
ν1
σ 2 for ν1 > 2 .
ν1 − 2 1
If the strategy of chossing a CGD such that E(θ|θn ) = θn is adopted (as discussed earlier; this implies that
the average of the moves is the current value) then σ12 needs to be selected so that
E σ 2 ν1 , σ12 =
This can be done by selecting σ12 =
ν1 −2 2
ν1 σn ,
since in that case,
Thus, the distribution
E σ 2 σn2 = σn2 .
E σ 2 σn2 =
ν1
σ 2 = σn2 .
ν1 − 2 1
ν1 − 2 2 σn
ν1
is the CGD chosen with ν1 being a “tuning” constant that can be varied to improve mixing of the chain.
q
σn2 ,
σ
2
≡ SI −χ2 ν1 ,
17
2
2. If X ∼ χ2 (ν), then Y = νXσ ∼ SI −χ2 (ν, σ 2 ). To generate a random variate σ 2 from the scaled-inverse
chi-squared distribution SI −χ2 (ν, σ 2 ), generate x from χ2 (ν) and set σ 2 = νσ 2 /x.
3. Because of the form of α(σn2 , σ∗2 ), it is convenient to compute it as exp (log(α)). This involves computing
the log posterior and log CGD densities each time through the loop in the M-H algorithm.
log(α) = log(posterior(σ∗2 )) + log(CGD(σ∗2 , σn2 )) − log(posterior(σn2 )) − log(CGD(σn2 , σ∗2 ))
4. Note that log(posterior) = log(prior) + log(likelihood) where
log(prior) = log [h(σ 2 |νp , σp2 )]
νp σp2
νp
+ 1) log(σ 2 ) −
2
2σ 2
2
log(likelihood) = log [`(σ |y)]
= c1 − (
= c2 −
n
n
1 X
log(σ 2 ) − 2
(yi − µ)2
2
2σ i=1
n
Y
i
h
1
1
√
exp − 2 (yi − µ)2 . Note that the constant c1 and c2 cancel out in computing
2σ
2πσ 2
i=1
log (α) above so need not be exactly determined.
since
`(σ 2 |y)
=
5. Also note that
log(CGD(σn2 , σ 2 )) = log [q(σn2 , σ 2 )]
ν1
ν1
(ν1 − 2)σn2
= c3 +
log(σn2 ) − ( + 1) log(σ 2 ) −
2
2
2σ 2
after some simplification. Again c3 cancels out in computing log acceptance ratio log(α) although it
depends on ν1 .
6. In the R functions supplied the arguments are in the order shown below:
generate.CGD (ν, σ 2 ): generate from SI −χ2 (ν, σ 2 )
log.prior (σ 2 , νp , σp2 ): compute log π(σ 2 )
log.lik (σ 2 , y, µ): compute log likelihood
log.post (σ 2 , y, µ, νp , σp2 ): compute log posterior
log.CGD (σn2 , ν1 , σ 2 ): compute log proposal density
MH.normal.variance (y, µ, νp , σp2 , σ02 , ν1 , nB , nM , nT , seed, output.file.prefix)
alpha(σn2 , σ∗2 , y, µ, νp , σp2 , ν1 ): compute acceptance probability
18
#-----------------------------------------------------------# R functions to do Metropolis-Hastings sampling
# for the NB10 data
#
# prior:
sigma2 ~ SI-chisq( nu.p, sigma2.p )
# data model: ( y_i | sigma2 ) ~IID N( mu, sigma2 ), i = 1, ..., n
#
#
#-----------------------------------------------------------MH.normal.variance = function( y, mu, nu.p, sigma2.p,
sigma2.0, nu.star, n.burnin, n.monitor, n.thin, seed ) {
# Main routine
sigma2.old = sigma2.0
R=0
set.seed( seed )
for ( i in 1:n.monitor) {
sigma2.star = generate.CGD( nu.star, ( nu.star - 2 ) * sigma2.old /
nu.star )
u = runif( 1 )
b = ( u <= alpha( sigma2.old, sigma2.star, y, mu, nu.p, sigma2.p,
nu.star ) )
sigma2.new = sigma2.star * b + sigma2.old * ( 1 - b )
if ( i > n.burnin ) R = R + b
if ( ( i > n.burnin ) & ( ( i - n.burnin ) %% n.thin == 0 ) )
write( c( ( i - n.burnin ) / n.thin, signif(sigma2.new, digits = 5
)),
file="nb10.output", ncol = 2, append = T )
}
return( R / (n.monitor-n.burnin) )
}
#-------------------------------------------------------generate.CGD = function( nu, sigma2 ) {
# candidate generating distribution
return( nu * sigma2 / rchisq( 1, nu ) )
}
#--------------------------------------------------------alpha = function( sigma2.old, sigma2.star, y, mu, nu.p, sigma2.p,
nu.1 ) {
# Acceptance probability calculation
return( min( 1, exp( log.post( sigma2.star, y, mu, nu.p, sigma2.p ) +
log.CGD( sigma2.star, nu.1, sigma2.old) - log.CGD( sigma2.old, nu.1,
sigma2.star) log.post(sigma2.old, y, mu, nu.p, sigma2.p ) ) ) )
}
#----------------------------------------------------------log.post = function( sigma2, y, mu, nu.p, sigma2.p ) {
# log( posterior ) calculation
return( log.lik( sigma2, y, mu ) + log.prior( sigma2, nu.p, sigma2.p
) )
}
#-----------------------------------------------------------log.lik = function( sigma2, y, mu ) {
# log( likelihood ) calculation
n = length( y )
return( ( - n / 2 ) * log( sigma2 ) - sum( ( y - mu )^2 )/( 2 *
sigma2 ) )
}
#-----------------------------------------------------------log.prior = function( sigma2 , nu.p, sigma2.p ) {
# log( prior ) calculation
return( ( -1 - nu.p / 2 ) * log( sigma2 ) - nu.p * sigma2.p /
( 2 * sigma2 ) )
}
#-----------------------------------------------------------log.CGD = function( sigma2.old, nu.1, sigma2 ) {
# log( candidate generating density ) calculation
return( ( nu.1 / 2 ) * log( sigma2.old ) - ( 1 + nu.1 / 2 ) *
log( sigma2 ) - ( nu.1 - 2 ) * sigma2.old/ ( 2 * sigma2 ) )
}
Gibbs Sampler
If π(·) is a multivariate target distribution e.g., π(x), then the entire vector x will be updated all at once by
generating the y from a proposal density q(x, y), using the Metropolis-Hastings algorithm. Instead, the updating
may be done componentwise, where the components of x may be of any dimension. For the purpose of discussion,
consider all components of x to be of single dimension i.e. x = (x1 , x2 , . . . , xk ). Each of these components are
then updated one by one sequentially in separate Metropolis-Hastings steps. For example, at the i th step, yi is
generated from the proposal density qi (xi , yi ) where qi depends on the current value of xi and may depend on
any of the other components of x, namely x−i = (xi , . . . , xi−1 , xi+1 , . . . , xk ), as well. The candidate yi is
accepted with acceptance probability
πi (yi ) qi (yi , xi )
αi (xi , yi ) = min
, 1
πi (xi ) qi (xi , yi )
If yi is accepted, set the ith component of xn , xn,i = yi ; otherwise set xn,i = xn,i . The remaining components of
xn are not changed in step i. This is repeated for i = 1, . . . , k, at end of which the entire vector x n would have
been updated.
The above is called a single component Metropolis-Hastings algorithm. Here π i (xi ), called the full conditional
distribution of xi , is the distribution of the ith component of x conditioning on all remaining components of x:
πi (xi ) = R
π(x)
.
π(x)dxi
Here we are using the result that a joint density (i.e. π(x)) is uniquely determined by the set of full conditionals
πi (xi ), i = 1, . . . , k.
A special single-component Metropolis-Hastings is the Gibbs sampler. For the Gibbs sampler, the proposal
distribution for updating the ith component of x is
qi (xi , yi ) = πi (yi ) ,
where πi (yi ) is the full conditional distribution of yi with respect to π(·). That is yi is generated from πi (yi ). If
qi (xi , yi ) above is substituted in the expression for αi (xi , yi ), it turns out to be equal to 1; i.e. Gibbs sampler
candidates are always accepted. Thus Gibbs sampling consists of sampling from full conditionals of the target
distribution.
Example 1:
Consider generating bivariate random variables from the density
f (x, y) =
n
x
!
y x+α−1 (1 − y)n−x+β−1
It can be shown that
f (x|y) ∝
n
x
!
for x = 0, 1, . . . , n
and 0 < y ≤ 1
y x (1 − y)n−x
i.e., X|(Y = y) ∼ Bin(n, y). Similarly
f (y|x) ∝ y x+α−1 (1 − y)n−x+β−1
19
i.e. Y |(X = x) ∼ Beta(x + α, n + β). The Gibbs sampler for generating bivariate samples from f (x, y) is then
for i = 1, . . . , n repeat
1. generate yi from Beta(xi−1 + α, n + β)
2. generate xi from Bin(n, yi )
3. return (xi , yi )
The stationary or equilibrium
distribution of pairs (xi , yi ) is f (x, y) given above. It can be shown that the
R
global balance condition p(x, y)π(x)dx = π(y) holds in this case.
Hierarchical Models
Suppose we have a data model f (y|θ) and a prior distribution of θ with density g(θ|λ), that depends on a parameter λ that is an unknown random variable. Let the distribution, called the hyperprior, of the hyperparameter
λ have density π(λ) . We wish to obtain the posterior p(θ|y). But, f (y|θ)g(θ|λ) ∝ posterior of θ given y and λ.
Thus
f (y, θ)
f (y, θ)
p(θ|y) = R
=
,
f (y|θ)g(θ)dθ
h(y)
where h(y) is the marginal distribution of y. Since the joint density,
f (y, θ, λ) = f (y|θ, λ) f (θ, λ)
= f (y|θ) g(θ|λ) π(λ).
and
f (y, θ) =
we have that
p(θ|y) =
Z
Z
f (y, θ, λ) dλ
f (y, θ, λ) dλ/h(y)
Z
f (y|θ)
∝
| {z }
g(θ|λ)π(λ) dλ
|
{z
}
(4)
model × (marginal) prior on θ.
If the posterior of λ, p(λ|y) is needed
p(λ|y) = f (y, λ)/h(y) =
∝
(3)
Z
|
Z
f (y, θ, λ) dθ/h(y)
f (y|θ)g(θ|λ)dθ · π(λ)
{z
mixed model
}
× prior on λ.
The prior g(θ|λ) just “mixes” the model f (y|θ) over the values of θ giving a mixed model independent of θ.
20
(5)
In summary, once you have determined which posterior distribution you need for inference, the rest is easy.
Go back to the joint density f (y, θ, λ) and integrate out the appropriate variables. When the posterior can
be obtained in closed form, then the prior is said to be a conjugate prior. Thus if it is known that a prior is a
conjugate then posterior can be obtained by inspection. The same applies for obtaining full conditionals. The
harder problem is to evaluate the integrals in closed form when the priors are not conjugate. See below for an
application of the Gibbs Sampler methods to solve a problem in heirarchical modelling.
Example 2:
Consider a random sample y = (y1 , . . . , yn ) from a normal density with mean µ and variance τ −1 . Suppose that
µ is subject to a normal prior with mean 0 and variance ω −1 and τ is subject to a gamma prior with shape
parameter α and scale parameter β. Given that the two priors are independent, the joint density of data and
parameters is
Pn
n+1 n
τ
ω 2
1
2
τ α−1 − βτ
(2π)− 2 τ 2 e− 2 i=1 (yi −µ) · ω 2 e− 2 µ ·
e .
Γ(α)β α
Gibbs sampling from the joint posterior distribution of µ and τ requires the conditional density of µ given y and
τ , say, h(µ|bf y, τ ), and the conditional density of τ given y and µ, say, g(τ |bf y, µ).
Because of the the choice of conjugate priors the conditional densities are easily obtained. Note then that
τ
h(µ|y, τ ) ∝ e− 2
τ
Pn
i=1
∝ e− 2 (−
∝ e−(
∝ e
(yi −µ)2 − ω
µ2
2
Pn
i=1
2µyi +nµ2 )− ω
µ2
2
nτ +ω 2
µ −τ
2
Pn
y µ)
i=1 i
nȳτ 2
1
)
(µ− nτ
−
+ω
2(nτ +ω)−1
This implies that µ given y is normally distributed with mean nτ ȳ/(ω + nτ ) and variance 1/(ω + nτ ), where ȳ
P
is the sample mean n1 ni=1 yi . Further,
n
τ
g(τ |y, µ) ∝ τ 2 e− 2
n
Pn
∝ τ 2 +α−1 e
i=1
−τ (
τ
(yi −µ)2 α−1 − β
τ
Pn
i=1
(yi −µ)2
2
e
+ β1 )
Thus, τ given y and µ is gamma distributed with shape parameter n/2 + α and scale parameter 1/(ns 2n /2 + 1/β),
P
where s2n is the sample variance n1 ni=1 (yi −µ)2 . The Gibbs sampling scheme is easily constructed using specified
values of ω, α, and β.
21
References
Chib, S., and Greenberg, E. (1995),“Understanding the Metropolis-Hastings Algorithm,” The American Statistician, 49, 327–335.
Gelfand, A. E., and Smith, A. F. M. (1990), “Sampling-Based Approaches to Calculating Marginal Densities,”
Journal of the American Statistical Association, 85, 398–409.
Gelman, A. (1992), “Iterative and Non-Iterative Simulation Algorithms,” in Computing Science and Statistics
(Interfact Proceedings), 24, 433–438.
Gelman, A., Carlin, D. B., Stern, H.S., and Rubin, D. B. (1995), Bayesian Data Analysis, Chapman &
Hall:London
Gelman, A., and Rubin, D. B. (1992), “Inference from Iterative Simulation Using Multiple Sequences” (with
discussion), Statistical Science, 7, 457–511.
Geman, S., and Geman, D. ((1984) “Stochastic Relaxation, Gibbs Distributions and the Bayesian Restoration
of Images,” IEEE Transactions on Pattern Analysis and Machine Intelligence, 6, 721–741.
Geweke, J. (1989), “Bayesian inference in econometric models using Monte Carlo integration,” Econometrica,
57, 1317-1340.
Gilks, W. R. , Richardson, S. and Spiegelhalter, D. J. [Ed.] (1996) Markov Chain Monte Carlo in Practice,
Chapman & Hall:London.
Hastings, W. K. (1970), “Monte Carlo Sampling Methods Using Markov Chains and Their Applications,”
Biometrika, 57, 97–109.
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H., and Teller, E. (1953), “Equations of State
Calculations by Fast Computing Machines,” Journal of Chemical Physics, 21, 1087–1092.
Smith, A. F. M., and Roberts, G. O. (1993), “Bayesian Computation via the Gibbs Sampler and Related
Markov Chain Monte Carlo Methods,” Journal of the Royal Statistical Society, Scr. B, 55, 3–24.
Tanner, M. A., and Wong, W. H. (1987), “The Calculation of Posterior Distributions by Data Augmentation,”
Journal of the American Statistical Association, 82, 528–549.
Tierney, L. (1994), “Markov Chains for Exploring Posterior Distributions” (with discussion), Annals of Statistics, 22, 1701–1762.
22