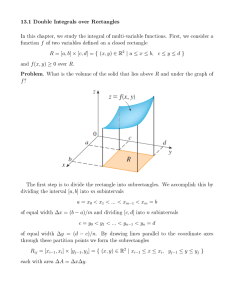
k-Means Clustering on Data with Incomplete Replications 1 Abstract Andy Lithio
advertisement

k-Means Clustering on Data with Incomplete Replications
Andy Lithio
May 9, 2013
1
Abstract
A flexible extension of the Hartigan-Wong k-means clustering algorithm for analysis of data where
observations have possibly unequal numbers of incomplete replications is proposed. Cluster initialization follows the suggestions of Hartigan and Wong. It is shown that the extension is equivalent
to the original Hartigan-Wong algorithm in the case of complete data with no replications. Computational and programming details are discussed, and the new algorithm is then applied to simulated
data with varying levels of censoring and complexity.
2
Introduction
The Hartigan-Wong k-means clustering algorithm is a routine for “[dividing] M points in N dimensions into K clusters so that the within-cluster sum of squares is minimized”. The algorithm
is not guaranteed to find a partition that achieves a global minimum for within-cluster sum of
squares, but obtains a local minimum where no movement of a point to a different cluster will
reduce the within-cluster sum of squares. Given appropriate initial values for the cluster means,
the Hartigan-Wong algorithm has been shown to perform well and be computationally efficient.
In this paper, we extend the Hartigan-Wong algorithm to the case where we have a set of observations, each consisting of at least one set of measurements, where each measurement is not
necessarily complete. We derive expressions for the reduction in within-cluster sum of squares
realized from transferring an observation out of a cluster and for the increase in within-cluster
sum of squares when transferring an observation into a cluster. These expressions are essential for
determining whether an observation should be moved to a different cluster. We then apply the
adapted algorithm to simulated datasets to investigate its efficiency and accuracy.
1
3
3
METHODS
Methods
Consider a set of observations X1 , ..., XN , where each Xi consists of ni measurements of dimension
n. We assume that each Xi has at least one recorded value in each dimension, but may contain
missing values. Define nij to be the number of recorded values for observation Xi in dimension
P
j. We define the input values M = N
i=1 ni , N to be the number of observations, and K to be
the number of clusters. Input data must be formatted as follows. Each line must contain one
measurement, with the recorded values listed in the first n columns, using nan to denote missing
values. All measurements from each Xi must be on consecutive lines. The (n + 1)th column should
contain an indexing variable from 1 to N identifying which Xi the observation corresponds to. The
data is read in and stored as the dat matrix.
Additionally, a k × (2n + 3) array of doubles named clust, a N × (n + 3) array of integers named
obs, and a N × 2n array of doubles named W OSS are created. Each row of clust corresponds to a
different cluster: the first n columns each contain one dimension of the current value of each cluster
center, the second n columns contain the number of recorded values in each dimension currently
assigned to that cluster, the (2n + 1)th column counts the number of Xi currently in each cluster,
the (2n + 2)th column is used to keep track of live clusters, and the final column contains the
within-cluster sum of squares for each cluster.
Each row of the array of integers obs corresponds to an observation, Xi . The first column contains
the number of measurements for each Xi , the second lists the cluster each Xi is currently assigned
to, with the third column essentially containing the next closest cluster to each Xi . Finally, the
final n columns list the number of recorded values in each dimension for each Xi . Each row of the
array W OSS also correspond to an Xi , but its first n columns contain the within-observation sum
of squares of that observation in each dimension, which will be used in the derivations of Section
3.1. The second n columns of W OSS contain the sum of the recorded values in each dimension of
the observation, which will be useful in updating cluster means.
The primary adjustment to the Hartigan-Wong algorithm required is to find the appropriate
quantities used to calculate the reduction in within-cluster sum of squares that would be realized
by removing an observation from its current cluster and the increase in within-cluster sum of
squares that would be realized when transferring an observation to another cluster. For data with
2
nL D(I,L)2
no replications and no missing values, Hartigan and Wong state these to be nLnD(I,L)
and
+1
nL −1 ,
L
respectively, where L is the cluster in question, I is the observation, nL is the number of observations
assigned to cluster L, and D(I, L)2 is the sum of squares between the observation I and cluster L.
The derivations for the reduction and increase in within-cluster sum of squares follow, transferring
observation Xi to or from cluster L, where L0 = L − Xi in the case of reduction and L0 = L ∪ Xi in
the case of increase in within-cluster sum of squares. Denote nLj as the number of recorded values
in dimension j assigned to cluster L, and recall nij is the number of recorded values for observation
Xi in dimension j. We will also use Yij to refer to the j th dimension
of a single measurement, so
P
that Xi = {Yij |Yij ∈ Xi }. For notational simplicity, denote X̄ij =
the j th dimension. Finally, we will denote µLj =
PM
i=1
I(Yij ∈L)Yij
nLj
i:Yij ∈Xi
Yij
, the center of Xi in
as the j th dimension of the center
of cluster L, and µL0 j as the j th dimension of the center of cluster L0 .
2
nij
3.1
3.1
Increase in Within-Cluster Sum of Squares
3
METHODS
Increase in Within-Cluster Sum of Squares
The increase
within-cluster sum of squares realized from transferring observation X1 to cluster
Pin
n
+
L is ∆+
=
j=1 ∆Lj , where
L
∆+
Lj
M
X
=
(Yij − µL0 j )2 I(Yi ∈ L0 ) − (Yij − µLj )2 I(Yi ∈ L)
=
i=1
M
X
2
(Yij − 2Yij µL0 j + µ2L0 j )I(Yi ∈ L0 ) − (Yij2 − 2Yij µLj + µ2Lj )I(Yi ∈ L)
i=1
=
X
Yij2
+
i:Yij ∈X1
=
X
M
X
(−2Yij µL0 j + µ2L0 j )I(Yi ∈ L0 ) − (−2Yij µLj + µ2Lj )I(Yi ∈ L)
i=1
Yij2 + −2(nLj + nij )µ2L0 j + (nLj + nij )µ2L0 j + 2nLj µ2Lj − nLj µ2Lj
i:Yij ∈X1
=
X
Yij2 + nLj µ2Lj − (nLj + nij )µ2L0 j
i:Yij ∈X1
=
X
Yij2
+
nLj µ2Lj
i:Yij ∈X1
=
X
Yij2
+
n2Lj µ2Lj + nLj nij µ2Lj
nLj + nij
i:Yij ∈X1
=
X
i:Yij ∈X1
P
0 2
( M
i=1 Yij ∗ I(Yi ∈ L ))
−
nLj + nij
+
−n2Lj µ2Lj − 2nLj µLj (
n
Lj
nij µ2Lj − 2nLj µLj
Yij2 +
nLj + nij
P
i:Yij ∈X1
nLj + nij
2
X
Yij
X
P
Yij ) − ( i:Yij ∈X1 Yij )2
i:Yij ∈X1
Yij −
nLj + nij
i:Yij ∈X1
2
X
=
(nLj + nij )
2
i:Yij ∈X1 Yij
P
nLj + nij
+
nLj
nij µ2Lj − 2nLj µLj
nLj + nij
nLj
nij µ2Lj − 2nLj µLj
=
nLj + nij
X
X
Yij +
i:Yij ∈X1
nij
Yij2 +
=
nLj
nLj + nij
X
nij
=
nLj
nLj + nij
i:Yij ∈X1
||Yij − µLj ||2 +
i:Yij ∈X1
nLj + nij
i:Yij ∈X1
X
X
Yij2 −
i:Yij ∈X1
||Yij − µLj ||2 +
i:Yij ∈X1
X
(Yij − X̄ij )2
i:Yij ∈X1
nLj + nij
3
i:Yij ∈X1
nLj + nij
2
nij
i:Yij ∈X1
nLj + nij
2
X
X
Yij2 −
Yij
i:Yij ∈X1
X
Yij
i:Yij ∈X1
X
Yij −
Yij
3.1
Increase in Within-Cluster Sum of Squares
=
nLj
nLj + nij
X
||Yij − µLj ||2 +
i:Yij ∈X1
3
nij
nLj + nij
X
METHODS
||Yij − X̄ij ||2
i:Yij ∈X1
The numerator of the term on the right is constant for each combination of observation and
dimension, so these are calculated and stored in the first n columns of the W OSS matrix after the
data is read in, then referenced when needed. The term on the right can be thought of as a weighted
corrected sum of squares for the measurements within Xi . The term on the left is reminiscent of the
quantity used by Hartigan-Wong, with the exception of incrementing by nij instead of 1, and having
to calculate it for each dimension instead of having a constant nLnL+1 multiplier for all dimensions.
The term on the left represents the distance between Xi and the current cluster center, and is
likely to have much more weight than the other term. The derivation for the reduction in withinclusterP
sum of squares when removing Xi from cluster L is similar, and results in the quantity
∆− = nj=1 ∆−
j , where
X
X
nLj
nij
||Yij − µLj ||2 −
||Yij − X̄ij ||2 .
∆−
j =
nLj − nij
nLj − nij
i:Yij ∈X1
i:Yij ∈X1
With ∆− and ∆+ defined, we now simply follow the Hartigan-Wong algorithm using observations
X1 , ..., XN , with ∆− and ∆+ as the reduction and increase in within-cluster sum of squares, respectively. To initialize the cluster centers, we adapt the suggestion of Hartigan and Wong. We first
calculate the overall mean for each dimension of the data, then put each observation in ascending
order by the distance from its mean to the overall mean. Then, for cluster L ∈ {1, 2, ..., K}, we
th
assign the mean of the 1 + (L−1)N
ordered observation (rounding to the nearest integer) to
K
be the the initial center. This ensures that no cluster will be empty after the initial assignment
of observations to clusters. However, the reader should note that this initialization method may
not be efficient or lead to accurate assignments, and further testing is required to determine a
recommended initialization. The remainder of the algorithm is outlined below, with comments on
how the included C program accomplishes each step.
Step 1: Initial Assignment For each observation, find the closest and second closest cluster centers,
as defined by the sum of squared errors, and record the index of each cluster in the second
and third columns of obs. Each observation is assigned to its closest cluster center.
Step 2: Update Cluster Centers Update the cluster centers to be the averages of all measurements
of observations assigned to them. At this point we also compute and store the number
of observations assigned to each cluster, as well as the number of recorded values in each
dimension for assigned to each cluster, and the within-cluster sum of squares. We track the
within-cluster sum of squares because it is convenient and useful for debugging, but adds
a very small computational burden. If desired, the within-cluster sum of squares can be
calculated only once upon exiting the algorithm.
Step 3: Live Set Initialization Put all clusters in the live set. In the included program, the (2n+2)th
column of clust is used to track which clusters are in the live set, where 0 indicates the cluster
is not in the live set, and any non-zero values indicates the cluster is in the live set. On this
step all entries in that column are given the value 2N + 1.
4
4
APPLICATION
Step 4: OPTRA Consider each observation I (I=1,...,N). If cluster L was updated in the last quicktransfer (QTRAN) stage, it will stay in the live set at least through the end of this stage.
Otherwise, remove a cluster from the live set if it has not been updated in the last N optimaltransfer (OPTRA steps). Let I be in cluster L1. If L is in the live set, do Step 4a, otherwise
do Step 4b.
Step 4a: Compute the minimum of ∆+
L over all clusters {L|L 6= L1}. Let L2 be the cluster that
has that minimum. If ∆+
≥
∆− , no transfers are necessary, but we record L2 as the
L2
next closest cluster to I. Otherwise, transfer I to L2, and record L1 as the next closest
cluster. Update the centers, within-cluster sum of squares, and counts of observations
of clusters L1 and L2. L1 and L2 are now in the live set, and we keep track of when the
clusters were last involved in a transfer by setting their live set columns to N + I.
Step 4b: Do Step 4a, but computing the minimum of ∆+
L only over the clusters in the live set.
Step 5: Exit Check Stop if the live set is empty. This will be the case if no transfers were made in
Step 4. Otherwise, proceed to Step 6.
Step 6: QTRAN Step Consider each observation I (I=1,...,N). Let L1 be the cluster I is assigned
to, and L2 be the cluster recorded as the next closest. We need not check I if both L1 and
−
L2 have not changed in the last N steps. If ∆+
L2 ≥ ∆ , no change is necessary. Otherwise,
switch L1 and L2, update the centers, and record their involvement in a transfer, and update
their live set columns to 2N + 1.
Step 7: Transfer Switch If no transfer has taken place in the last N steps (the count variable is
greater than or equal to N ), return to Step 4. Otherwise, return to Step 6.
4
Application
Using R scripts provided by Dr. Maitra, we performed simulations on the performance of this
adapted algorithm on data with varying levels of censoring and clustering complexity. Each data
set had n = 2 dimensions, k = 5 clusters, N = 1000 observations, and the same vectors of ni so
that M = 4547. The first measurement of each observation was left as complete, and the remaining
measurements were censored at rates varying from 0 to 0.8. At each censoring level, 100 data
sets were generated, and the Adjusted Rand Index (ARI) of the assignments made by the adapted
Hartigan-Wong and the true clusters was calculated and recorded. This process was repeated for
different complexities of clustering, as determined by M axOmega, ranging from 0.001 to 0.75. In
the table below, we report the mean of the 100 simulations at each combination of censoring and
M axOmega under the REP method. For comparison, we also run the original Hartigan-Wong
routine using just the first measurement of each observation on each data set, using both the initialization described above (the ORD method) and R’s default initialization of choosing 5 points
at random (the RAND method). Keep in mind that the censoring rate does not affect the ORD
and RAND methods, but they are included for comparison to the REP method.
5
5
Table 1: Mean ARI
MaxOmega=
Censoring Rate REP
0
0.755
0.1
0.776
0.2
0.784
0.4
0.785
0.6
0.788
0.8
0.817
0.001
ORD RAND
0.778 0.770
0.786 0.770
0.753 0.762
0.817 0.786
0.760 0.793
0.824 0.779
REP
0.705
0.670
0.684
0.677
0.670
0.671
0.25
ORD RAND
0.624 0.653
0.622 0.623
0.609 0.626
0.629 0.598
0.622 0.611
0.632 0.618
REP
0.226
0.230
0.225
0.207
0.294
0.212
DISCUSSION
0.75
ORD RAND
0.190 0.184
0.194 0.193
0.200 0.199
0.181 0.178
0.174 0.166
0.200 0.200
The apparent lack of a trend (or even the increasing trend) in mean ARI under REP as the
censoring rate increases is unexpected. It may indicate that there is a bug in the code or simulation
methods, but also may simply mean that not much is gained from the replicates (remember, the
first measurement of each observation in this simulation was complete). To compare, note that
under M axOmega = 0.001, there is nearly no difference between the REP method and the others.
However, as M axOmega increases, we see the mean ARI under REP uniformly higher than under
the other methods. Comparing the ORD and RAND methods, we also see no indication that one
initialization is superior to the other, but a more detailed comparison would be required to fully
investigate this matter.
5
Discussion
We have extended the Hartigan-Wong k-means clustering algorithm to operate on data with incomplete replicates by deriving new terms for the increase and decrease in within-cluster sum of squares
realized by moving an observation into or out of a cluster. We limited testing of the algorithm to
varying levels of censoring and clustering complexity, but it may also be interesting to investigate
performance for different numbers of clusters, replicates, or dimensions. A more in-depth comparison of different initialization methods might also be of interest, especially between choosing the
means k observations at random to function as the initial centers and the method outlined above.
There is also room for further work on the C implementation of the adapted algorithm. There
is potential to reduce the number of computations in the included code by storing ∆− for each
observation, and only updating when necessary (I attempted to include this, but ran out of time).
Finally, the included program assumes that each observation has at least one recorded value in each
dimension. Theoretically, this should not be necessary, but a program allowing such data is still
being debugged and tested.
6