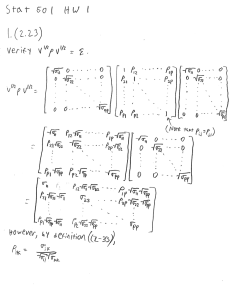
HW1 Solution #1.6) 5
advertisement

HW1 Solution 3.0 #1.6) (a) Plot the marginal dot diagrams for all the variables. 10 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 5 6 7 8 9 10 ● 2.0 ● ● ● ● ●● ● ● ● 40 2 3 4 5 6 7 8 2 4 6 count 10 5 4 ● ● ● ● ● ● ● 3 ● ● ● ● ● ● ● ● 2 ● ● ● ● ● ● ● ● ● 1 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 25 20 15 10 2.5 3.0 ● 20 4 ● ●●●●● ● ●●●●●●●●● ●● ●●●●●●●●●●● ●● 5 ●● 10 15 ● ●●● 20 25 O3 ● ● ● ● ● ● ● ● ● 3.5 ● 3 count 5 ● ●● 2 ● NO2 2.0 ● 1 ● 15 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● NO ● 10 ● ● ● ● ● ● ● ● ● ● 100 ● 6 6 ● ● ● ● 5 80 ● 14 ● ● ● ● ● ● ● ● 15 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 10 ●● ●●●●●● ●●●●●●●●●●●●●● solar radiation ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 5 count ● ●●● ● 60 CO count ●● 1.5 ● ● 1.0 ● ● count ● ● 6 2 4 count 8 ● wind count ● 2.5 ● 5 ●● ● 4.0 ● 4.5 5.0 HS Figure 1: Dot plots of the variables in the air pollution dataset. (b) Construct the x̄, Sn , and R arrays, and interpret the entries in R. 1 Wind 7.50 x̄ Solar 73.86 Table 1: Wind 2.50 -2.78 -0.38 -0.46 -0.59 -2.23 0.17 Wind Solar CO NO NO2 O3 HC Table 2: Wind Solar CO NO NO2 O3 HC CO 4.55 NO 2.19 NO2 10.05 O3 9.40 HC 3.10 The sample means of variables. Solar -2.78 300.52 3.91 -1.39 6.76 30.79 0.62 CO -0.38 3.91 1.52 0.67 2.31 2.82 0.14 NO -0.46 -1.39 0.67 1.18 1.09 -0.81 0.18 NO2 -0.59 6.76 2.31 1.09 11.36 3.13 1.04 O3 -2.23 30.79 2.82 -0.81 3.13 30.98 0.59 HC 0.17 0.62 0.14 0.18 1.04 0.59 0.48 The sample variance-covariance matrix of the variables. Wind 1.00 -0.10 -0.19 -0.27 -0.11 -0.25 0.16 Table 3: Solar -0.10 1.00 0.18 -0.07 0.12 0.32 0.05 CO -0.19 0.18 1.00 0.50 0.56 0.41 0.17 NO -0.27 -0.07 0.50 1.00 0.30 -0.13 0.23 NO2 -0.11 0.12 0.56 0.30 1.00 0.17 0.45 O3 -0.25 0.32 0.41 -0.13 0.17 1.00 0.15 HC 0.16 0.05 0.17 0.23 0.45 0.15 1.00 The sample correlation matrix of the variables. The majority of the variables have only weak linear associations, with correlations close to zero. The pollutants are mostly positively correlated with each other. Wind is negative correlated with pollutants, while solar radiation is positively correlated with pollutants. 2 Windy and Sunny Windy and Not Sunny 1 4 6 10 14 15 7 16 22 17 18 19 23 26 28 20 21 30 31 35 42 36 Not Windy and Sunny 2 3 5 9 12 25 27 37 38 39 40 Figure 2: Not Windy and Not Sunny 8 11 13 24 29 32 33 34 41 Star plots of the air pollution variables. To investigate if there is an effect on air pollution with wind and the sun, we can divide the wind and solar radiation variable in half by the median and then make the star plots in Figure 2. From the stars, we can see that solar radiation have some effects on the air pollution. When it is sunny, most pollutants are on a relative low level. And within each group, the patterns are quite different. So there are a fair amount of variation in each of the four groups, as there were days with very little pollution in each group, as well as days with quite a bit of air pollution. 3 #1.17) In this dataset the first three are measured in seconds, while the last four are measured in minutes. x̄ 100m 11.62 200m 23.64 Table 4: 100m 200m 400m 800m 1500m 3000m Marathon Table 5: 100m 200m 400m 800m 1500m 3000m Marathon 100m 0.20 0.48 1.01 0.04 0.11 0.28 9.44 400m 53.41 800m 2.08 1500m 4.33 3000m 9.45 Marathon 173.25 The sample means for the track record. 200m 0.48 1.23 2.55 0.09 0.26 0.65 23.18 400m 1.01 2.55 7.17 0.26 0.70 1.72 57.49 800m 0.04 0.09 0.26 0.01 0.03 0.08 2.57 1500m 0.11 0.26 0.70 0.03 0.11 0.27 8.88 3000m 0.28 0.65 1.72 0.08 0.27 0.68 22.57 Marathon 9.44 23.18 57.49 2.57 8.88 22.57 925.96 The sample variance covariance matrix for the track record variables. 100m 1.00 0.95 0.83 0.73 0.73 0.74 0.69 Table 6: 200m 0.95 1.00 0.86 0.72 0.70 0.71 0.69 400m 0.83 0.86 1.00 0.90 0.79 0.78 0.71 800m 0.73 0.72 0.90 1.00 0.90 0.86 0.78 1500m 0.73 0.70 0.79 0.90 1.00 0.97 0.88 3000m 0.74 0.71 0.78 0.86 0.97 1.00 0.90 Marathon 0.69 0.69 0.71 0.78 0.88 0.90 1.00 The sample correlation matrix for the track record variables. All the seven variables are strongly positively correlated. And the correlations tend to be larger when distances are close to each other.For example he correlation between 100m and 200m is 0.95, while the correlation between 100m and marathon is 0.69. This makes sense, since runners are good at races of similar length. 4 #1.18) 1 100m 8.62 Table 7: 100m 200m 400m 800m 1500m 3000m Marathon 200m 8.48 400m 7.51 800m 6.44 1500m 5.81 3000m 5.33 Marathon 4.15 The sample mean track records measured in meters/second. 100m 0.11 0.12 0.10 0.08 0.10 0.10 0.13 200m 0.12 0.15 0.13 0.09 0.11 0.12 0.16 400m 0.10 0.13 0.14 0.11 0.12 0.12 0.15 800m 0.08 0.09 0.11 0.11 0.12 0.12 0.15 1500m 0.10 0.11 0.12 0.12 0.16 0.16 0.20 3000m 0.10 0.12 0.12 0.12 0.16 0.17 0.21 Marathon 0.13 0.16 0.15 0.15 0.20 0.21 0.32 Table 8: The sample variance covariance matrix of track records measured in meters/second. 100m 200m 400m 800m 1500m 3000m Marathon 100m 1.00 0.95 0.84 0.73 0.74 0.75 0.72 200m 0.95 1.00 0.86 0.73 0.72 0.72 0.71 400m 0.84 0.86 1.00 0.90 0.80 0.78 0.71 800m 0.73 0.73 0.90 1.00 0.92 0.87 0.79 1500m 0.74 0.72 0.80 0.92 1.00 0.96 0.86 3000m 0.75 0.72 0.78 0.87 0.96 1.00 0.89 Marathon 0.72 0.71 0.71 0.79 0.86 0.89 1.00 Table 9: The sample correlation matrix of track records measured in meters/second. The results are similar with those I obtained in Exercise 1.17. All have strong, positive linear relationships with each other, and races that are closer together in distance have a stronger relationship. The differences in correlation are slightly less maybe because all the races were measured in the same units now. Compute the sample variance-covariance matrix (call it S). Obtain the spectral decomposition (also called the eigenvalue decomposition: use eigen if you are using R) of the variance covariance matrix. Next post-multiply the observation matrix (call it X) with P. Plot the pairwise scatter plots of the first three columns. 5 −7.4 −7.0 −6.6 −6.2 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ● ● ● ● ●● ● ● ● ● ● ● ● ● var 2 ● ● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ●● ● ● ● ● ●●● ● ● ● ● ● ● ●● ● ●● ● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ● ● ● ●● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● −1.4 −7.4 ● ● ●● ● ●● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ● ● ● ● ● ● ● var 3 ●● −1.8 −6.2 −6.6 ●● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ● ● ●● ● ● ● ●● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ●● ● ●● ●● ● ● ● ● ●● −7.0 ● ●● ● −17 var 1 −18 ● −15 ● ● −16 ● −14 ● ● ● ● ● −2.2 ● ● −18 −17 Figure 3: −16 ● −15 −14 −2.2 −1.8 −1.4 pairwise scatter plots of the first three columns of Y=X*P. From the pairwise scatter plots, we could not find any obvious relationships among these three variables. The first two columns may have some negative relationship while the last two columns have a slightly positive relationship. #1.26 (a) x̄ Breed 4.38 SaleP 1742.43 Table 10: YearlingHT 50.52 FFBody 995.95 PctFFBody 70.88 Frame 6.32 The sample means of the variables in the bulls dataset. 6 Back.fat 0.20 SaleHT 54.13 Salewt 1555.29 Breed SaleP YearlingHT FFBody PctFF Frame Back.fat SaleHT Salewt Breed 9.68 -434.74 2.83 117.83 4.80 1.25 -0.17 3.04 46.94 Table 11: SaleP -434.74 388133.66 456.47 5890.60 -229.47 276.42 15.44 486.97 25645.89 YearlingHT 2.83 456.47 3.00 100.13 2.96 1.51 -0.05 2.98 82.81 FFBody 117.83 5890.60 100.13 8594.34 209.50 51.95 -1.40 129.94 6680.31 PctFF 4.80 -229.47 2.96 209.50 10.69 1.46 -0.14 3.41 83.93 Frame 1.25 276.42 1.51 51.95 1.46 0.86 -0.02 1.49 44.32 Back.fat -0.17 15.44 -0.05 -1.40 -0.14 -0.02 0.01 -0.05 2.41 SaleHT 3.04 486.97 2.98 129.94 3.41 1.49 -0.05 4.02 147.29 Salewt 46.94 25645.89 82.81 6680.31 83.93 44.32 2.41 147.29 16850.66 The sample variance covariance matrix of the variables in the bulls dataset. Breed SaleP YearlingHT FFBody PctFFBody Frame Back.fat SaleHT Salewt Breed 1.00 -0.22 0.52 0.41 0.47 0.43 -0.62 0.49 0.12 Table 12: SaleP -0.22 1.00 0.42 0.10 -0.11 0.48 0.28 0.39 0.32 YearlingHT 0.52 0.42 1.00 0.62 0.52 0.94 -0.34 0.86 0.37 FFBody 0.41 0.10 0.62 1.00 0.69 0.60 -0.17 0.70 0.56 PctFFBody 0.47 -0.11 0.52 0.69 1.00 0.48 -0.49 0.52 0.20 Frame 0.43 0.48 0.94 0.60 0.48 1.00 -0.26 0.80 0.37 Back.fat -0.62 0.28 -0.34 -0.17 -0.49 -0.26 1.00 -0.28 0.21 The sample correlation matrix of the variables in the bulls dataset. Only a few variables(Frame, Yearling height, and Sale Height) have strong relationships with each other. I do not think the breeds are well separated in this system since all the correlations between breed and other variables are not strong. The best potential variable to distinguish between breeds is back fat, which has the strongest linear relationship with breed. (b) I did not find any obvious outliers from Figure 4. From the three dimensional plot, we can observe that most bulls with breed 8 (Simental) have less back fat and larger frame. And the values of back fat and frame in breed 1 (Angus) are more spread out. 7 SaleHT 0.49 0.39 0.86 0.70 0.52 0.80 -0.28 1.00 0.57 Salewt 0.12 0.32 0.37 0.56 0.20 0.37 0.21 0.57 1.00 Figure 4: A three dimensional plot. (c) This time the points are more closely clustered, so it is more clearly to separate these three breeds. Bulls with breed 8 (Simental) have higher fat free body weight and higher sale height. And the values of fat free body weight and sale height in breed 1 (Angus) are more spread out. 8 Figure 5: A three dimensional plot. 9 #2.20) 0.526 A = PΛ P = 0.851 0.526 A−1/2 = P Λ−1/2 P 0 = 0.851 1.376 0.325 0.761 A1/2 A−1/2 = 0.325 1.701 −0.145 1/2 1/2 0 2 1 A= 1 3 −0.851 1.902 0 0.526 0.851 1.376 0.325 = 0.526 0 1.176 −0.851 0.526 0.325 1.701 −0.851 0.526 0 0.526 0.851 0.761 −0.145 = 0.526 0 0.851 −0.851 0.526 −0.145 0.616 −0.145 0.761 −0.145 1.376 0.325 1 0 = = =I 0.616 −0.145 0.616 0.325 1.701 0 1 # 2.23) √ V 1/2 ρV 1/2 = σ11 √ σ22 .. . √ 1 ρ12 .. . ρ12 1 .. . ... ... .. . √ σ11 ρ1p ρ2p .. . σpp ρ1p ρ2p . . . 1 √ √ √ √ σ11 ρ12 σ11 σ22 . . . ρ1p σ11 σpp √ √ ρ12 √σ11 √σ22 σ22 . . . ρ2p σ22 σpp = .. .. .. .. . . . . √ √ √ √ ρ1p σ11 σpp ρ2p σ22 σpp . . . σpp σ11 σ12 . . . σ1p σ12 σ22 . . . σ2p = . .. .. = Σ .. .. . . . σ1p σ2p . . . σpp 10 √ σ22 .. . √ σpp