Modeling changes in gene expression in neurodegeneration in mice Lalita Devadas
advertisement

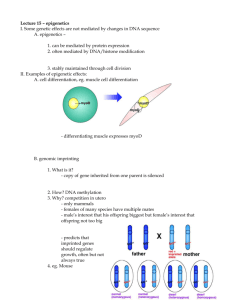
Introduction Biological Background Building Models Results Moving Forward Modeling changes in gene expression in neurodegeneration in mice Fourth Annual Primes MIT Conference Lalita Devadas Mentor: Angela Yen May 18, 2014 Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 1/18 Introduction Biological Background Building Models Results Moving Forward Outline 1 Biological Background 2 Building Models 3 Results 4 Moving Forward Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 2/18 Introduction Biological Background Building Models Results Moving Forward Outline 1 Biological Background 2 Building Models 3 Results 4 Moving Forward Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 3/18 Introduction Biological Background Building Models Results Moving Forward Gene Expression Central dogma of molecular biology Regulated by genetic (ACTG) and epigenetic factors Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 4/18 Introduction Biological Background Building Models Results Moving Forward Epigenetics Histone modifications DNA Enviroment Epigenetic factors are context that affect gene expression Histone Modifications Chemical changes to histone protein core or protruding tail Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 5/18 Introduction Biological Background Building Models Results Moving Forward Experimental Data Neurodegeneration in mice Control Mouse Neurodegenerative Mouse Gene Expression Data Gene Expression Data Change in Gene Expression Data Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 6/18 Introduction Biological Background Building Models Results Moving Forward Outline 1 Biological Background 2 Building Models 3 Results 4 Moving Forward Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 7/18 Introduction Biological Background Building Models Results Moving Forward Types of Models Random Forest Random Forest model Returns value based on set of values determined by a group of decision trees Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 8/18 Introduction Biological Background Building Models Results Moving Forward Types of Models Linear Model Linear model Finds a linear correlation between predictors and response Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 9/18 Introduction Biological Background Building Models Results Moving Forward Two-Step Model Classification and Regression Chromatin Mark Data (∼3000 genes) Classification More Expressed than control (∼400 genes) Similarly Expressed (∼2200 genes) Less Expressed than control (∼400 genes) Regression Regression Regression Predicted Expression Change Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 10/18 Introduction Biological Background Building Models Results Moving Forward Outline 1 Biological Background 2 Building Models 3 Results 4 Moving Forward Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 11/18 Introduction Biological Background Building Models Results Moving Forward Final Model Train on half of data, test on other half Classification step: Random Forest model Regression step: Linear models Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 12/18 Introduction Biological Background Building Models Results Moving Forward Results of Two-Step Model Classification Graph Accuracy of Classification Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 13/18 Introduction Biological Background Building Models Results Moving Forward Results of Two-Step Model Classification Values Expression Sensitivity Specificity More than control .199 .043 Less than control .063 .043 Same as control .971 .128 Sensitivity how good the model is at predicting if a data point belongs in a certain class Specificity how good the model is at predicting if a data point doesn’t belong in a certain class Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 14/18 Introduction Biological Background Building Models Results Moving Forward Results of Two-Step Model Regression Graph Accuracy of Regression: r 2 ≈ 0.18 Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 15/18 Introduction Biological Background Building Models Results Moving Forward Outline 1 Biological Background 2 Building Models 3 Results 4 Moving Forward Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 16/18 Introduction Biological Background Building Models Results Moving Forward Next Steps Reprocess data (to improve our predictive power) Use different data (possibly Roadmap data) Create R package (for cross validation) Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 17/18 Introduction Biological Background Building Models Results Moving Forward Acknowledgements I would like to thank: My mentor, Angela Yen Prof. Manolis Kellis Andreas Pfenning Prof. Li-Huei Tsai and Elizabeth Gjoneska (Experimental collaborators) PRIMES program My family Lalita Devadas — Modeling changes in gene expression in neurodegeneration in mice 18/18