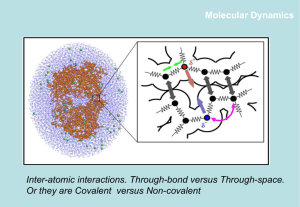
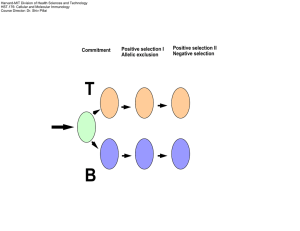
Overview of Immunology and Molecular Dynamics
advertisement
Brief overview of Immunology and Molecular Dynamics Denise Chac 10 October 2014 Immunology • The study of the human immune system Lymphocytes B Cells • Membrane bound antibody • Activation: bind to pathogen and triggered by helper T cells • MHC II proteins present antigen peptide on cell surface • Differentiates – Effector Cells – Memory Cells T Cells • T Cell Receptors (TCR) • Helper T Cells – Presents CD4 – Recognize MHC class II – Activation: release of cytokines • Cytotoxic T Cells – Presents CD8 – Recognizes MHC class I – Destroys infected/cancerous cells B Cell T-Cells Viruses HIV/AIDS Previous Studies • The breadth of epitope recognition within specific regions of HIV antigen peptides contribute to anti-viral efficiency of CD8 T-cell response [Geldmacher et al. 2007] • The altering of antigen peptides on MHC to give a more ‘protruding’ surface topology may increase TCR repertoire diversity [Turner et al. 2005] Method: Molecular Dynamics • VMD – Visual Molecular Dynamics – Program to visualize large biomolecular systems in 3-D graphics • Molecular Dynamics - NAMD • MODELLER – Homology studies – Python based homology modeling software used for modeling proteins Methods: NAMD • Parallel molecular dynamics code designed for highperformance simulation of large biomolecular systems • Based on Charm++ but can be compatible with AMBER, CHARMM, and X-PLOR • Need: – Protein Data Bank – PDB • Atomic coordinates • Velocities – Protein Structure File – PSF • Structural information – Force Field Parameter File • Mathematical potential of atoms in the system – Configuration File • Tells how NAMD will run the simulation Methods: NAMD – the solvent • Put protein in water to resemble the cellular environment • How? – A water sphere in surrounding vacuum • Without periodic boundary conditions – A water box • With periodic boundary conditions Questions: • “epitope enhancement” • Difference between R5 and X4 HIV • TCR flexibility [Martinez-Hackert et al. 2006] References • • • • • • Martinez-Hackert, E., Anikeeva, N., Kalams, S.A., Walker, B.D., Hendrickson, W.A., Sykulev, Y., 2006. Structural basis for degenerate recognition of natural HIV peptide variants by cytotoxic lymphocytes. Journal of Biological Chemistry 281, 20205–20212. Geldmacher, C., Currier, J.R., Herrmann, E., Haule, A., Kuta, E., McCutchan, F., Njovu, L., Geis, S., Hoffmann, O., Maboko, L., Williamson, C., Birx, D., Meyerhans, A., Cox, J., Hoelscher, M., 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. Journal of Virology 81, 2440–2448. Turner, S.J., Kedzierska, K., Komodromou, H., La Gruta, N.L., Dunstone, M.A., Webb, A.I., Webby, R., Walden, H., Xie, W., McCluskey, J., Purcell, A.W., Rossjohn, J., Doherty, P.C., 2005. Lack of prominent peptide–major histocompatibility com- plex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nature Immunology 6, 382–389. Park, M., Park, S.Y., Miller, K.R., Collins, E.J., Lee, H.Y., 2013. Accurate Structure prediction of peptide-MHC complexes for identifying highly immunogenic antigens. Molecular Immunology 56, 81-90. Alberts, B., Johnson, A., Lewis, J. et al. 2002. Molecular Biology of the Cell. 4th Edition. New York: Garland Science. Janeway, C., Murphy, K., Travers, P., Walport, M. 2008. Janeway’s Immunobiology. 8th Edition. New York: Garland Science .