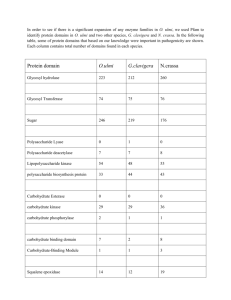
Table 1. The Oxomolybdenum Enzymes
advertisement

Molybdenum 1900 – discovered in plant ashes 1930 - azotobacter (N2 fixer) growth requirement for Mo 1940 – established as essential micronutrient for plants 1950 - [Mo] affected rat XO activity; [Mo]~ [flavin] Mo associated with aldehyde oxidase in rabbit livers 1960 - Mo crucial to N-cycle Mo The only 4d metal essential to Life S Table 1. The Oxomolybdenum Enzymes The Xanthine Oxidase Family (LMoOS-Possessing Enzymes) enzyme source subunits cofactora xanthine oxidase cow’s milkb R2 MPT xanthine dehydrogenase chicken liverc R2 MPT rat liverd R2 MPT Micrococcus lactyliticuse Drosophila melanogasterf R2 MPT Chlamydomonas reinhardtiig humanh R2 (MPT) aldehyde oxidase rabbit liveri R2 (MPT) humanj R2 (MPT) cowk R2 (MPT) aldehyde oxidoreductase (dehydrogenase) Desulfovibrio gigasl R2 MCD Acetobacter polyoxogenesm formate dehydrogenase Alcaligenes eutrophusn Râçä Methylosinus trichosporumo R2â2ç2ä2 CO dehydrogenase (oxidoreductase) Pseudomonas carboxydovoransp R2â2ç2 Pseudomonas carboxydoflavaq R2â2ç2 MCD Oligotropha carboxidovoransr R2â2ç2 MCD quinoline-2-oxidoreductase Pseudomonas putida s R2â2ç2 MCD Rhodococcus sp. B1t R2â2ç2 MCD Comamonas testosteroni 63u R2â2ç2 MCD isoquinoline 1-oxidoreductase Pseudomonas diminutav Râ MCD quinoline-4-carboxylate-2-oxidoreductase Agrobacterium sp. 1Bw R2â2ç2 MCD quinaldine-4-oxidoreductase Arthrobacter sp.x R2â2ç2 MCD quinaldic acid 4-oxidoreductase Serratia marcescensy Pseudomonas sp. AK-2z Râ nicotinic acid hydroxylase (dehydrogenase) Clostridium barkeriaa,ab R2 Bacillus niaciniac R2â2ç2 Arthrobacter oxidansad Râç 6-hydroxynicotinate hydroxylase Bacillus niaciniac Râç nicotine dehydrogenase Arthrobacter oxidansad Râç Arthrobacter nicotinovoransae picolinate hydroxylase Arthrobacter picolinophilusaf R2â2ç2 MCD (2R)-hydroxycarboxylate oxidoreductase Proteus vulgarisag (R. Hille, Chem. Rev. 1996) The Sulfite Oxidase Family (LMoO2-Possessing Enzymes) sulfite oxidase bovine liverah R2 MPT chicken liverai R2 MPT rat liveraj R2 MPT humanak R2 MPT Thiobacillus novellusal R nitrate reductase (assimilatory) Neurospora crassaam R2 MPT spinachan R2 MPT Chlorella vulgarisao R4 MPT The DMSO Reductase Family (L2MoX-Possessing Enzymes) DMSO reductase Rhodobacter sphaeroidesap R MGD Rhodobacter capsulatus aq R MGD Escherichia coli ar Râç MGD biotin-S-oxide reductase Escherichia colias trimethylamine-N-oxide reductase Escherichia coliat R2 nitrate reductase (dissimilatory) Escherichia coli (NarGHI)au Râç MGD Escherichia coli (NarZYV)av Râç MGD Escherichia coli (FdoGHI)aw Râç MGD Paracoccus denitrificans (NapABCD)ax Râçä Haloferax volcaniiay formate dehydrogenase Escherichia coli (FdhF)az R MGD Escherichia coli (FdnGHI)ba Râç MGD Escherichia coli (FdoGHI)bb Râç MGD Methanobacterium formicicumbc (XdS) Râç MGD Wollinella succinogenesbd Râç polysulfide reductase Wolinella succinogenesbe Râç MGD arsenite oxidase Alcaligenes faecalisbf R MCD formylmethanofuran dehydrogenase) Râçä(ú) MGD, MAD, MHD Methanosarcina barkeribh MGD Unclassified Molybdenum-Containing Enzymes pyridoxal oxidase Drosophila melanogaster chlorate reductase Proteus mirabilis tetrathionite reductase Proteus mirabilis xanthine dehydrogenase Clostridium sp pyruvate:ferredoxin oxidoreductase Archaeoglobus fulgidus pyrogallol transhydroxylase Pelobacter acidigallici 2-furoyl-CoA dehydrogenase Pseudomonas putida Locating the Molybdenum Cofactor All plants require the molybdenum enzyme All mammals require molybdenum enzymes Nitrate Reductase Sulfite Oxidase NO3- + 2H+ + 2e- SO32- + H2O NO2- + H2O SO42- + 2H+ + 2e- Mo(4+) + X-O + 2H+ Mo(6+) + X + H2O NO2- Nitrate reductase N2 nitrogenase Nitrogen- cycle NO3- NH3 polysulfide reductase SH2 CH4 Sx Carbon- cycle Sulfur- cycle CO2 CH3CO2- (CH3)2S (CH3)2SO DMSO reductase Table 1. The Oxomolybdenum Enzymes (R. Hille, Chem. Rev. 1996) The Xanthine Oxidase Family (LMoOS-type) enzyme xanthine oxidase xanthine dehydrogenase source cofactor cow’s milk MPT chicken liver MPT rat liver MPT Drosophila melanogaster MPT human MPT) aldehyde oxidase rabbit liver MPT human MPT cow MPT aldehyde oxidoreductase (dehydrogenase) Desulfovibrio gigas MCD formate dehydrogenase Alcaligenes eutrophus CO dehydrogenase (oxidoreductase) Pseudomonas carboxydovorans quinoline-2-oxidoreductase Pseudomonas putida MCD isoquinoline 1-oxidoreductase Pseudomonas diminuta MCD quinoline-4-carboxylate-2-oxidoreductase Agrobacterium sp MCD quinaldine-4-oxidoreductase Arthrobacter sp. MCD quinaldic acid 4-oxidoreductase Serratia marcescens Pseudomonas sp. AK nicotinic acid hydroxylase (dehydrogenase) Clostridium barkeriaa, 6-hydroxynicotinate hydroxylase Bacillus niaciniac nicotine dehydrogenase Arthrobacter oxidansa The Sulfite Oxidase Family (LMoO2-type) enzyme source sulfite oxidase bovine liver chicken liver rat liver human Thiobacillus novellus nitrate reductase (assimilatory) Neurospora crassa spinach Chlorella vulgari The DMSO Reductase Family (L2MoX-type) DMSO reductase Rhodobacter sphaeroides biotin-S-oxide reductase Escherichia colia trimethylamine-N-oxide reductase Escherichia colia cofactor MPT MPT MPT MPT MPT MPT MPT MGD nitrate reductase (dissimilatory) Escherichia coli (NarGHI) MGD formate dehydrogenase Escherichia coli (FdhF) MGD polysulfide reductase Wolinella succinogene MGD arsenite oxidase Alcaligenes faecalisb MCD formylmethanofuran dehydrogenase) MGD, MAD, MHD Unclassified Molybdenum-Containing Enzymes pyridoxal oxidase Drosophila melanogaster chlorate reductase Proteus mirabilis tetrathionite reductase Proteus mirabilis xanthine dehydrogenase Clostridium sp. pyruvate:ferredoxin oxidoreductase Archaeoglobus fulgidus pyrogallol transhydroxylase Pelobacter acidigallici 2-furoyl-CoA dehydrogenase Pseudomonas putida The Generic Molybdenum Cofactor MAD MGD MCD MPT There’s not only one, but a family of Moco’s Why molybdopterin? dithiolene Mo pterin Table 1. The Oxomolybdenum Enzymes (R. Hille, Chem. Rev. 1996) The Xanthine Oxidase Family (LMoOS-type) The Sulfite Oxidase Family (LMoO2-type) The DMSO Reductase Family (L2MoX-type) Used to be classed with XO b/c structural similarity. Unique Mo enzyme with a 2nd metal, Cu Table 1. The Oxomolybdenum Enzymes (R. Hille, Chem. Rev. 1996) The Xanthine Oxidase Family (LMoOS-type) The Sulfite Oxidase Family (LMoO2-type) Xanthine Oxidase/Dehydrogenase - in mammalian milk but >50% de-Mo - in liver: purine metabolism (gout) - implicated in heart damage (ROS form’n) Nitrate Reductase - in all plants - assimilatory form (plants, fungi, algae) The DMSO Reductase Family (L2MoX-type) - all bacterial or from archea - many in respiration using specific e- acceptors - recall DMSOR in cloud formation …. - simplest Mo-enzymes: w/o other cofactors Sulfite Oxidase - most critical for humans - detoxification of sulfite - also has role in S-metabolism, from Cys Met SO32- SO42- Table 1. The Oxomolybdenum Enzymes (R. Hille, Chem. Rev. 1996) The Xanthine Oxidase Family (LMoOS-type) - in mammalian milk but >50% de-Mo - in liver: purine metabolism (gout) - implicated in heart damage (ROS form’n) The Sulfite Oxidase Family (LMoO2-type) Nitrate Reductase - in all plants - assimilatory form (plants, fungi, algae) NO3- + 2H+ + 2e- NO2- + H2O The DMSO Reductase Family (L2MoX-type) - all bacterial or from archea - many in respiration using specific e- acceptors - recall DMSOR in cloud formation …. - simplest Mo-enzymes: w/o other cofactors Sulfite Oxidase - most critical for humans - detoxification of sulfite - also has role in S-metabolism, from Cys Met SO32- SO42SO32- + H2O SO42- + 2H+ + 2e- Root nodules formed by Rhizobium bacteria living symbiotically through nitrogen fixation. Tomato plants with and without molybdenum available. Tobacco plants (Arabidopsis thaliana) Nitrate Nitrite Proteins nitrate Healthy (wild type) All plants require Assimilatory Nitrate Reductase Sick (mutant) nitrite Introduction Repairing the Molybdenum Cofactor Baby Z Cured of Rare Disease in 3 Days University of Arizona, Tucson, October 2010 (Southern Health/AFP/Getty Images) Orphan Drug Treatment Used Only on Mice to Get Hearing Before FDA Baby Z had a one in a million By SUSAN chance of developing rare9, 2009 DONALDSON JAMES, aNov. metabolic disorder called molybdenum cofactor deficiency and zero chance of avoiding the inevitable death sentence that comes with it. Australian girl had acofactor, Worldwide, there are only about 50The cases of molybdenum seemingly normal in May or sulfite oxidase deficiency, mostly in Europe and inbirth the United 2008 but,ofwithin hours, she began States, according to the National Institutes Health. havingismultiple as Molybdenum, like other organic metals, essentialseizures for the -human many asmolecule 10 an hour as sulfite body. Its cofactor is a small, complicated that--acts as a Why the correct oxidation state matters MRI of brain of deceased baby with Sulfite Oxidase Deficiency MRI of healthy brain The baby died because this reaction didn’t happen: SO32- + Sulfite S4+ H2O ---> SO42- + 2H+ + Sulfate S6+ 2e- The baby has a genetic defect in the enzyme that catalyzes this reaction. Babies with this genetic disease die within hours. The enzyme is Sulfite Oxidase. You have it in your liver. Review the terms: Oxidation half reaction Reduction half reaction SO32- + H2O S4+ in Sulfite ---> SO42- + 2H+ + 2eS6+ in Sulfate Mo6+ + 2e- ---> Mo4+ Net redox reaction SO32- + H2O + Mo6+ ---> SO42- + 2H+ + Mo4+ Is this reaction spontaneous? Caroline Kisker Würzburg, Germany Protein crystallographer X-ray structure of chicken liver Sulfite Oxidase Kisker, Enemark Moco locked into position by H-bonds to pterin A catalytic cycle for how Mo oxidizes SO32coupled oxygen electron proton transfer S atom + SO3 2- O transfer +6 Mo S O O S O - H+, - e- O S O O Mo+5 OH O - H+, - e- SO42- S S O O S S a S Mo S S O S Mo+4 O H H S + H2O Mo+4 O O Mo+4 S O O Water mediated OAT Similar cycles can be devised for Sulfite Oxidase and Nitrate Reductase SO32- + H2O Sulfite S4+ NO3- + Nitrate N5+ ---> SO42- + 2H+ + Sulfate S6+ 2H+ + 2e- 2e- ---> NO2- + H2O Nitrite N3+ Environmental impact of DMSOR and the smell of the ocean DMS CH3SO3- cloud nucleation sites hn, photo-oxidation DMS (CH3)2S(CH2)2CO2from algae DMSO Reductase DMSO DMSOR Structure: Controversy #2 The first Mo enzyme X-ray structure: DMSO Reductase Group Meeting Bryn Mawr College, October 2010 Doug Rees, 1996 Doug Rees, Ca Protein crystal SURPRISE!!!! • 2 molydopterin ligands! • nucleoside termini on pterin • very long Mo-S bonds DMSOR Structure: Controversy #2 The first look at molybdopterin was on a tungsten enzyme! Hyperthermophilic TungstonEnzyme, Aldehyde Ferredoxin Oxidoreductase Group Meeting Bryn Mawr College, October 2010 Doug Rees et al., Science,1995 SURPRISE!!!! • not the molydopterin ligand! • is that pyran ring actually right??? DMSOR Structure: Controversy #2 Group Meeting Bryn Mawr College, October 2010 Will the real active site structure in DMSO Reductase please stand up? (S J N Burgmayer, in Progress in Inorganic Chemistry, 2004) And the answer was: Hermann Schindelin, Würzburg, Germany Protein crystallographer DMSOR Structure: Controversy #2 Group Meeting Bryn Mawr College, October 2010 1.3 Å X-ray Structure in DMSO Reductase (Schindelin) Active form What does it mean? There are 2 superimposed structures. (only one is inactive!) Inactive form