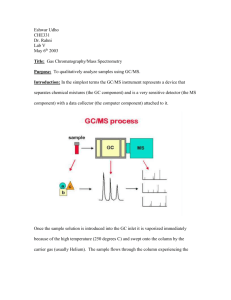
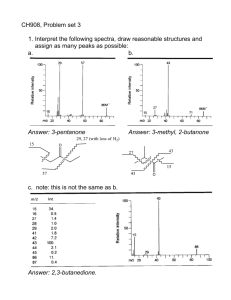
lecture 3, MS
advertisement

Mass Spectrometry A truly interdisciplinary and versatile analytical method MS is used • for the characterization of molecules ranging from small inorganic and organic molecules to polymers and proteins. With MS we might be able to determine the molecular weight of a molecule ion, the elemental composition of a molecule ion, and the presence of certain functional groups. • for mechanistic studies of gas-phase (ion) chemistry. This is also relevant for a better understanding of the chemistry in our atmosphere and in space. • as a mass detector coupled to a GC, HPLC, capillary electrophoresis (CE), and thermo-gravimetric analysis. • as an integrated mass detector in many high vacuum systems. UofW seminar speaker January 2002 The Gas-phase ion chemistry of cluster ions Dr Paul M. Mayer Assistant Professor Chemistry Department University of Ottawa History of MS Instrumentation http://masspec.scripps.edu/information/history/ History of MS Applications 1899 1956 Early Mass Spectrometry; Identifying Organic Compounds with MS; 1964 1966 1974 1990 1991 GC/MS Peptide Sequencing Extraterrestrial Mass Spectrometry Protein Structure Non-Covalent Interactions with ESI 1992 1993 1993 1996 Low Level Peptide Analysis Oligonucleotide Sequencing Protein Mass Mapping/Fingerprinting MS of a Virus 1999 1999 Desorption/Ionization on Silicon Isotope-Coded Affinity Tags MS Celebrities Ion Chemistry Francis William Aston (1877 - 1945) Cambridge University, Great Britain; Nobel Prize in Chemistry 1922 1st MS Joseph John Thomson (1856 - 1940) Cambridge University, Great Britain; Nobel Prize in Physics 1906 ESI of Biomolecules John B. Fenn (1917) Virginia Commonwealth University, Richmond, Virginia Ion Trap Technique Wolfgang Paul (1913 - 1993) University of Bonn, Germany; Nobel Prize in Physics 1989 Peptide Sequencing using MS Klaus Biemann (1926) MALDI MIT, Cambridge, Franz Hillenkamp (1936) Massachusetts University of Münster, Germany Mechanisms and Applications R. Graham Cooks (1941) Department of Chemistry, Purdue University West Lafayette, Indiana Fragmentation Mechanisms Fred W. McLafferty (1923) Cornell University Ithaca, New York Mechanism of MALDI & ESI Michael Karas (1952) University of Frankfurt, Germany Basic Concepts A mass spectrometer is an instrument that produces ions and separates them in the gas phase according to their mass-to-charge ratio (m/z). Today a wide variety of mass spectrometers is available but all of these share the capability to assign mass-to-charge values to ions, although the principles of operation and the types of experiments that can be done on these instruments differ greatly. Basically, a mass spectrometric analysis can be envisioned to be made up of the following steps: Sample Introduction → Ionization → Mass Analysis → Ion Detection/Data Analysis Samples may be introduced in gas, liquid or solid states. In the latter two cases volatilization must be accomplished either prior to, or accompanying ionization. Many ionization techniques are available to produce charged molecules in the gas phase, ranging from simple electron (impact) ionization (EI) and chemical ionization (CI) to a variety of desorption ionization techniques with acronyms such as FAB (fast atom bombardment), PD (plasma desorption), ES (electrospray) and MALD (matrix assisted laser desorption). The following pages are in part from http://ms.mc.vanderbilt.edu/tutorials/ms/ Sample Introduction Mass spectrometers are operated at reduced pressure in order to prevent collisions of ions with residual gas molecules in the analyzer during the flight from the ion source to the detector. The vacuum should be such that the mean free path length of an ion, i.e., the average distance an ion travels before colliding with another gas molecule, is longer than the distance from the source to the detector. For example, the mean free path length of an ion is approximately one meter at a pressure of 5x10-5 torr, i.e. about twice the length of a quadrupole instrument. Thus, the introduction of a sample into a mass spectrometer usually requires crossing of a rather large pressure drop, and several means have been devised to accomplish this. Gas samples may be directly connected to the instrument and metered into the instrument via a needle valve. Liquid and solid samples can be introduced through a septum inlet or a vacuum-lock system. However, when connecting continuous introduction techniques like gas chromatography (GC), high performance liquid chromatography (HPLC) or capillary electrophoresis (CE), special interfacing becomes imperative to prevent excessive gas load. Organic Structural Spectroscopy by Lambert, Shurvell, Lightner Ionization The deciding criteria are often the following: 1. Physical state of the sample 2. Volatility and thermal stability of the sample 3. Type of information sought Comparison • • • • EI, CI, and DI are suitable for high resolution MS; EI works well only for thermally stable and volatile samples; CI, SI, and DI cause much less fragmentation (normally no radicals are formed); DI and SI must be combined with tandem mass detection (MS-MS) to extract more structural information from fragmentation; Organic Structural Spectroscopy by Lambert, Shurvell, Lightner Electron Impact Ionization (EI) purely physical processes! The ionization energies of many compounds are on the order of 7-14 eV but electron energies of 70 eV are often chosen for EI-MS to achieve higher signal intensities and to avoid changes in the mass spectrum with small changes in electron energy. The energy domain: 1 J (kg m2 s-2) = 6.24145 x 1018 eV 1 J mol-1 = 1.03641 x 10-5 eV energies of chemical bonds are typically between 100 and 600 kJ mol-1 (C-H = 435 kJ mol-1; C-O = 356 kJ mol-1, C-N = 305 kJ mol-1), which are equivalent to 1-6 eV The time domain: a 70 eV electron has a velocity of about 5 x 108 cm s-1 and transits a molecule of 1 nm length in 2 x 10-16 s, while a typical bond vibration requires >10-12 s; thus, the molecular conformation remains unchanged as the electronic excitations occurs (Franck-Condon principle) The degree of fragmentation depends on the internal energy deposition in the molecular ion and on the resistance of the molecular structure to bond cleavage (fragmentation). Schematic representation of an electron ionization ion source. M represents neutral molecules; e-, electrons; M+· , the molecular ion; F+, fragment ions; Vacc, accelerating voltage; and MS, the mass spectrometer analyzer. Schematic representation of an electron ionization ion source. about every 1/1000 molecule is ionized the sample is heated up until a sufficient vapour pressure is obtained sample pressure in the ion source is about 10-5 torr only cations Advantages & Disadvantages of EI-MS Organic Structural Spectroscopy by Lambert, Shurvell, Lightner Contrasting degrees of fragmentation depending on the chemical structure Radical cations of aromatic and unsaturated conjugated hydrocarbons resist fragmentation more than radical cations of saturated hydrocarbons Lowering the electron beam energy to approximately 15 eV does not necessarily result in the observation of a molecular ion. CH3C=O++ •OCH2CH2CH2CH3 CH2=CHCH2CH3+ + CH3COOH It does, however, change the probability of different fragmentation pathways. Here, the H-rearrangement wins at the expense of single-bond cleavage. The Molecular Ion in EI-MS Index of hydrogen deficiency (degree of unsaturation) The index of hydrogen deficiency is the number of pairs of hydrogen atoms that must be removed from the corresponding saturated formula to produce the molecular formula of the compound of interest. The index is the sum of the number of rings, double-bonds, and twice the number of triple bonds. Compounds can contain C, H, N, O, halogen, and S. Index = tetravalent (C, Si) - ½ monovalent (H, halogen) + ½ trivalent (N, P) + 1 Bivalent atoms such as O and S do not contribute. The nitrogen rule Molecular ions containing odd numbers of nitrogen must have an odd number m/z. Example The compound with the molecular formula C7H7NO has an index of ?? What are possible structures? Note that a benzene ring counts for an index of 4, 3 double bonds and one ring! The isotopic signature – recognition of elements