DNA replication to translation
advertisement

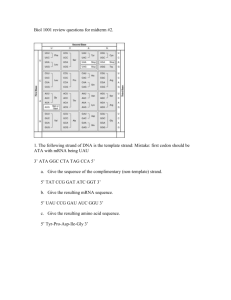
DNA DNA • must carry information • must be replicatable (inheritance) • must be changeable (mutation) DNA DNA structure deoxyribonucleic acid - two directional polynucleotide strands in a double helix A brief digression for terminology: Carbon molecules in rings are numbered…. 5C O 4 C1 C C 3 C 2 two directional polynucleotide strands in double helix start with a ribose sugar… two directional polynucleotide strands in double helix start with a ribose sugar… remove an oxygen at carbon 2’…. two directional polynucleotide strands in double helix start with a ribose sugar… remove an oxygen at carbon 2’…. add a phosphate group at 5’ side add a nitrogenous base at 1’ side = a nucleotide two directional polynucleotide strands in double helix A nucleotide, or base Bases = purines (adenine, guanine) and pyrimidines (cytosine, thymine) two directional polynucleotide strands in double helix nucleotides are linked in chains with a phosphodiester bond free ends of chain will have 5’ phosphate at one end, 3’ hydroxyl at the other end 5’ end phosphodiester bond 3’ end two directional polynucleotide strands in double helix nucleotides are linked in chains with a phosphodiester bond free ends of chain will have 5’ phosphate at one end, 3’ hydroxyl at the other end 5’ end 3’ end two directional polynucleotide strands in double helix Hydrogen bonds two directional polynucleotide strands in double helix Two strands pair up, nucleotides linked with hydrogen bonds adenosine pairs with thymine cytosine pairs with guanine two directional polynucleotide strands in double helix Two strands pair up, nucleotides linked with hydrogen bonds adenosine pairs with thymine cytosine pairs with guanine - abbreviated as “base pairs” two directional polynucleotide strands in double helix Strands have polarity - 5'-hydroxyl group of first nucleotide at one end, 3'-hydroxyl group at other end (5’ to 3’ strand) Strands run antiparallel: (5' -> 3') ATGGAATTCTCGCTC (3' <- 5') TACCTTAAGAGCGAG DNA replication: two strands are both available as templates for new strand result is doubling (2 complete new double helices) DNA replication: is semiconservative always occurs in 5’ to 3’ direction DNA replication: occurs at multiple replication forks (bubbles) along the DNA strand Important: there are several DNA polymerases involved in replication DNA polymerases have a proof-reading and editing function (exonuclease activity) TRANSCRIPTION Consider: if all DNA was actively used: - most mutations would be lethal - there would be no ‘raw material’ for evolutionary change - what would happen to genes de-activated by mutation? In fact, many errors and duplications leave ‘extra’ DNA Consider: If there is excess DNA, it may be - only between genes - also interspersed within genes Consider: If there is excess DNA, it may be - only between genes - also interspersed within genes Consider: Not all gene products are required simultaneously; needs for proteins change or differ - during development (e.g., milk digesting enzymes) - over time (e.g., digestive enzymes) - among organs (e.g., liver enzymes not used in muscle) - in response to stimuli (e.g., melanin, adrenalin) therefore regulation of gene activity is needed Transcription: Uses RNA as an intermediary - to assemble genes - to transmit the right information when/where it is needed (regulation) Transcription: Uses RNA as an intermediary - to assemble genes - to transmit the right information when/where it is needed (regulation) RNA is ribonucleic acid - has uracil instead of thymine - sugar is ribose instead of deoxyribose There are three types of RNA: mRNA: messenger RNA – carries the code for a gene rRNA: ribosomal RNA – used to construct ribosomes tRNA: transfer RNA – short adapters to carry amino acid and its anti-codon DNA strand (double, helical) - permanent (5' -> 3') ATGGAATTCTCGCTC (coding, sense strand) (3' <- 5') TACCTTAAGAGCGAG (template, antisense strand) DNA strand (double, helical) - permanent (5' -> 3') ATGGAATTCTCGCTC (coding, sense strand) (3' <- 5') TACCTTAAGAGCGAG (template, antisense strand) mRNA strand (single, linear) – temporary, as needed (5' -> 3') AUGGAAUUCUCGCUC (from template strand) DNA strand (double, helical) - permanent (5' -> 3') ATGGAATTCTCGCTC (coding, sense strand) (3' <- 5') TACCTTAAGAGCGAG (template, antisense strand) mRNA strand (single, linear) – temporary, as needed (5' -> 3') AUGGAAUUCUCGCUC (from template strand) note: by taking information from the template (antisense) strand of DNA, mRNA becomes the coding sequence DNA strand (double, helical) - permanent (5' -> 3') ATGGAATTCTCGCTC (coding, sense strand) (3' <- 5') TACCTTAAGAGCGAG (template, antisense strand) mRNA strand (single, linear) – temporary, as needed (5' -> 3') AUGGAAUUCUCGCUC (from template strand) protein sequence (single, with 1, 2, 3, 4 structure) Met-Glu-Phe-Ser-Leu... Gene structure promoter region: immediately upstream (5’ end) of its gene Steps in transcription: 1. initiation RNA polymerase recognizes and binds to promoter sequence - these contain TATAAA and TTGACA or CCAAT codes Steps in transcription: 1. initiation RNA polymerase recognizes and binds to promoter sequence - these contain TATAAA and TTGACA or CCAAT codes 2. elongation - similar to DNA replication - only one strand (template) is used Steps in transcription: 1. initiation RNA polymerase recognizes and binds to promoter sequence - these contain TATAAA and TTGACA or CCAAT codes 2. elongation - similar to DNA replication - only one strand (template) is used 3. termination - transcription keeps going for 1000-2000 bases beyond end of ‘gene’ After transcription: RNA processing capping polyadenylation intron removal promoter elements UTR= untranslated region TRANSLATION: The Genetic Code The genetic code DNA and RNA have 4 types of bases proteins are composed of amino acids, of which there are 20 - so how do 4 bases encode 20 amino acids? The genetic code “words” with a single base allow no combinations (4 words) “words” with two bases allow 16 combinations (42) “words” with three bases allow 64 combinations (43) = more than enough combinations for 20 amino acids The genetic code • composed of nucleotide triplets (codons) mRNA protein sequence AUG GAA UUC UCG CUC Met Glu Phe Ser Leu The genetic code • composed of nucleotide triplets (codons) • non-overlapping mRNA protein sequence NOT AUG GAA UUC UCG CUC Met Glu Phe Ser Leu AUGGAAUUCUCGCUC The genetic code • composed of nucleotide triplets (codons) • non-overlapping • unambiguous – each codon only specifies one amino acid • degenerate – most amino acids specified by several codons third position first position second position Reading frame must be uniquely specified: theredfoxatethehotdog t her edf oxa tet heh otd og th ere dfo xat eth eho tdo g the red fox ate the hot dog start codon Reading frame must be uniquely specified: mRNA code begins with start codon (AUG) protein is constructed along open reading frame translation stops at stop codon (UAA, UAG, or UGA) (only in frame: sequence out of frame does not work) Reading frame must be uniquely specified: mRNA code begins with start codon (AUG) protein is constructed along open reading frame translation stops at stop codon (UAA, UAG, or UGA) (only in frame: sequence out of frame does not work) 5’ 3’ GUCCCGUGAUGCCGAGUUGGAGUAAGUAACCU met pro ser trp ser lys stop The genetic code • composed of nucleotide triplets (codons) • non-overlapping • unambiguous • degenerate • nearly universal – except for portions of mitochondrial DNA and a few procaryotes TRANSLATION: assembling proteins Three types of RNA: mRNA: messenger RNA – carries the code for a gene 5’ 3’ GUCCCGUGAUGCCGAGUUGGAGUAGAUAACCU Three types of RNA: mRNA: messenger RNA – carries the code for a gene rRNA: ribosomal RNA – used to construct ribosomes - four types, used to make two-unit ribsome (30 S) (60 S) Three types of RNA: mRNA: messenger RNA – carries the code for a gene rRNA: ribosomal RNA – used to construct ribosomes tRNA: transfer RNA – short adapters to carry amino acid and its anti-codon anticodon Steps in translation: 1. initiation ribosomal subunits recognize, bind to 5’ cap on mRNA initiator tRNA (with UAC anticodon) binds to AUG start codon Steps in translation: 1. initiation 2. elongation next tRNA pairs with its codon peptidyl transferase 1. catalyzes formation of peptide bond between amino acids Steps in translation: 1. initiation 2. elongation next tRNA pairs with its codon peptidyl transferase 1. catalyzes formation of peptide bond between amino acids 2. breaks amino acid bond with previous tRNA ribosome shifts over one codon Steps in translation: 1. initiation 2. elongation 3. termination stop codon is recognized, bound to by release factor, polypeptide is freed Protein structure primary: amino acid sequence secondary: helix or pleated sheet, held with hydrogen bonds tertiary: collapsed molecule with internal bonds quaternary: protein subunits combine to form functional protein Protein structure quaternary: protein subunits combine to form functional protein subunits may be from same gene, or different may need two (dimers), three (trimers), or more Protein function enzymes – catalyze chemical reactions; most common proteins usually have active sites (tertiary structure) that mediate function structural proteins collagen, keratin transporters hemoglobin contractile – tissue and muscle movement actin, myosin intercellular communication insulin, other hormones Fig. 9-20