7 MBFiehn_Seattle_October_2010_Part_2
advertisement

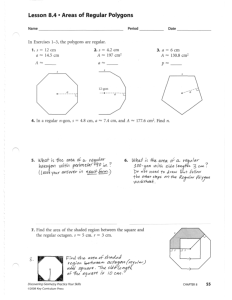
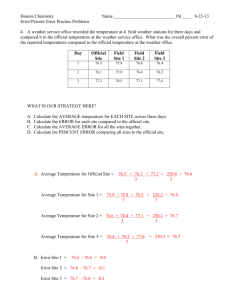
(1) Can we identify metabolites or metabolic pathways that are associated with breast cancer clinical parameters? Partial Least Square (multivariate stats) Score scatterplot (t1 vs. t2) Standard deviation of t1: 4.992 Standard deviation of t2: 7.089 15.0 grade2 12.5 grade1 grade1 10.0 grade3 7.5 5.0 2.5 0.0 t2 Results Alex-CIS-GCTOF MS w/ BinBase: 470 detected compounds 161 known metabolites, 309 without identified structure. breast adipose grade2 -2.5 -5.0 -7.5 -10.0 -12.5 grade3 -15.0 -17.5 -20.0 -12.5 -10.0 -7.5 -5.0 -2.5 t1 0.0 2.5 5.0 7.5 10.0 60 Grade n1 23 tumors show differential up-regulation in Pentose Phosphate Pathway, nucleotides, amino acids, arachidonic acid (but not free fatty acids, gln) 50 40 grade 1 Results 30 grade 2 grade 3 20 10 0 E+P+H- E+P+H+ E+P-H- E+P-H+ production of NADPH, ribose units E-P+H- E-P+H+ E-P-H- E-P-H+ production of DNA, RNA Grade n1 23 tumors show differential up-regulation in Pentose Phosphate Pathway, nucleotides, amino acids, arachidonic acid (but not free fatty acids, gln) Categ. Box & Whisker Plot: beta-alanine Categ. Box & Whisker Plot: glutamic acid 14000 6E5 12000 5E5 glu 10000 b-ala beta-alanine glutamic acid 4E5 3E5 2E5 2000 n 1 normal 2 3 grade 1 2 Categ. Box & Whisker Plot: malate 34000 3 grade n 1 2 3 grade 0 Mean Mean±SE normal 1 2 3 Mean±1.96*SE Categ. Box & Whisker Plot: citric acid grade Mean Mean±SE Mean±1.96*SE 55000 32000 30000 28000 26000 malate 50000 45000 citrate 24000 22000 lactate 40000 citric acid 20000 18000 malate 16000 14000 35000 30000 12000 10000 25000 8000 6000 4000 2000 20000 n 1 2 3 grade 15000 1 Plot: arachidonic 2 3 Categ. normal Box & Whisker acid 18000 16000 Amino acids n 1 Mean Mean±SE normal Mean±1.96*SE 2 3 grade 1 2 3 grade grade C20:4 2HOglutarate 14000 arachidonic acid Results 6000 4000 1E5 0 8000 Amino acids 12000 10000 8000 6000 4000 2000 n 1 normal 1 2 3 grade 2 3 Mean Mean±SE Mean±1.96*SE n 1 IDH1? 2 3 grade Mean Mean±SE Mean±1.96*SE Results Can we validate biomarkers in a fully independent study? Cohort 2 (113 patients) Matej Orešič et al. (subm.) Results Can we validate lipidomic biomarkers in a fully independent study? Cohort 2 (113 patients) Orešič et al. (subm) Results Can we validate lipidomic biomarkers in a fully independent study? Immunohistochemistry on tissue slides. Methods Can we get biochemical maps of differential regulation for hormone receptor status? (1) Up to 20% of the identified metabolites lack enzyme annotations. (2) Metabolic endpoint metabolites accumulate more than intermediates. ‘Comprehensive maps’ do not work. Methods Can we get biochemical maps of differential regulation for hormone receptor status? (1) Up to 20% of the identified metabolites lack enzyme annotations. (2) Metabolic endpoint metabolites accumulate more than intermediates. ‘Comprehensive maps’ do not work. Methods Chemical and biochemical mapping of metabolomic results Results Grade 1 tumors vs normal Red nodes: up-regulated in tumors at p<0.05, size ~ x-fold Blue nodes: down-regulated in tumors at p<0.05, size~x-fold Red edges: enzyme substrate/product in KEGGRpair DB Green edges: chemical similarity > 0.7 Hormone receptor @ grade 3 60 % of patients Methods 50 single neg. ER+ PR+ HER2- 40 grade 1 30 grade 2 grade 3 20 triple neg. ER- PRHER2- 10 0 E+P+H- E+P+H+ E+P-H- Estrogen positive E+P-H+ E-P+H- E-P+H+ E-P-H- Estrogen negative E-P-H+ Metabolic phenotype of triple negative tumors Results Patients without Estrogen, Herceptin neu2, Progesteron receptor Breast Cancer: Grade 3 ER- PR- HER- (n=27) Vs ER+PR+ HER- (n=19) Red : metabolite is enriched in triple negative Blue : metabolite is down regulated in triple negative Hormone receptor @ grade 3 60 % of patients Methods 50 40 grade 1 30 grade 2 grade 3 20 triple neg. ER- PRHER2- 10 double neg. ER- PRHER2+ 0 E+P+H- E+P+H+ E+P-H- Estrogen positive E+P-H+ E-P+H- E-P+H+ E-P-H- Estrogen negative E-P-H+ Results Effect of HER2 status on metabolic phenotype of triple negative tumors Breast Cancer: Grade 3 Red : metabolite is enriched in triple negative Hormone receptor @ grade 2 60 % of patients Methods 50 40 30 double pos. ER+ PR+ HER2- grade 1 grade 2 grade 3 20 10 triple pos. ER+ PR+ HER2+ 0 E+P+H- E+P+H+ E+P-H- Estrogen positive E+P-H+ E-P+H- E-P+H+ E-P-H- Estrogen negative E-P-H+