Regulation of Gene expression
advertisement
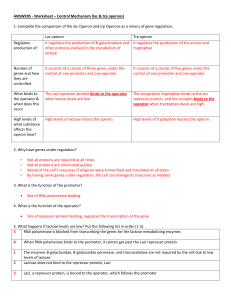
Regulation of Gene expression by E. Börje Lindström This learning object has been funded by the European Commissions FP6 BioMinE project Introduction • Biosynthetic reactions consume energy: • Available energy is limited in Nature: • The environment is important: Sophisticated control mechanisms in bacteria Production of as much cell material per energy as possible - the nutrient in the medium is used first - rapid and drastic changes in the nutrients - reversible control reactions needed • Two types of model systems: - Biosynthetic - Catabolic Biosynthetic reactions Tryptophan is chosen as a model system: - Tryptophan is an essential amino acid - Tryptophan is missing in some plant proteins - of industrial importance • The bacterial cells are controlling the biosynthesis of tryptophan in three ways: - feedback inhibition - end product repression - attenuation Biosynthetic reactions, cont. • Feedback inhibition: - The biosynthesis of tryptophan occurs in several steps: E1 antranilic acid E2 E3 C E4 E5 tryptophan Chorismate + glutamine B D Mechanism: - enzyme E1 (the first enzyme) is an allosteric protein with - a binding site for for the substrate - a binding site for the effectors (inhibitor = try) • E1 + try [E1-try]-complex that is inactive • the complete biosynthesis of try is stopped Biosynthetic reactions, cont. • End product repression (EPR): - In spite of ’end product inhibition’ - loss of energy due to enzymes E2-E5 are still synthesized - another regulation is needed - end product repression Biosynthetic reactions, cont. Mechanism: P O att E1 E2 E3 E4 E5 E1 – E5 = structural genes for the enzymes E1-E5. P = promoter; O = operator att = attenuator Initiation of mRNA synthesis • RNA polymerase binds to P • The repressor is an allosteric protein - inactive without tryptophan (does not bind to the operator) • tryptophan acts as co-repressor -binds to the repressor - makes the repressor active • The repressor binds to O Blocks the RNA polymerase movement Biosynthetic reactions, cont. • Attenuator region: - barrier for the RNA polymerase 1) + try the polymerase removed from the DNA 2) - try the polymerase continues into the structural genes • EPR inhibits all enzymes in tryptophan biosynthesis save energy - however, a slow total inhibition – does not effect already existing enzymes - high specificity – only the tryptophan operon is effected Biosynthetic reactions, cont. Biosynthetic reactions, cont. Biosynthetic reactions, cont. Biosynthetic reactions, cont. Catabolic reactions • Catabolic systems are inducible • The inducer is the available carbon/energy source • Model system – lactose operon in E. coli R P O lacZ lacY lacA • Where: - gene R : repressor protein – active without the inducer - blocks mRNA polymerase - gene lacZ : b-galactosidase – splits lactose into glycose + galactose - gene lacY: permease – transport lactose into the cell - no attenuator sequence in catabolic systems Catabolic reactions, cont. • Mechanism: + lactose: - transported into the cell transformed into allo-lactose (inducer) - allo-lactose + repressor [allo-lactose-repressor]- complex inactive - RNA polymerase starts transcription of lactose operon - b-galactosidase is produced break down of lactose - lactose: -[allo-lactose-repressor]- complex disintegrate - the repressor binds to O and blocks further transcription of the operon Catabolic reactions, cont. Catabolic reactions, cont. Catabolic repression (glucoseeffect) • Works in bacteria and other prokaryotes (here in E. Coli K12) • Diauxi: - growth on two energy sources glucose + lactose - two-step growth curve Log OD Growth on lactose lactose Growth on glucose time glucose Catabolic repression (glucoseeffect) • Mechanism: -cAMP an important substance - required for initiation of transcription of many inducible systems - global regulation - glucose present [cAMP] (decreases) - CAP (katabolite activator protein) an allosteric protein - [cAMP-CAP]-complex binds to the promoter promotes transcription -production of b-galactosidase -1) lactose present - 2) [cAMP-CAP]-complex present Catabolic repression (glucoseeffect), cont. • + glucose: - no [cAMP-CAP]-complex - no transcription of lactose operon - no b-galactosidase production • - glucose: - [cAMP-CAP]-complex present - transcription of lactose operon - b-galactosidase production - brake down of lactose Catabolic repression (glucoseeffect), cont. • Conclusions: - Katabolite repression – a very useful function in bacteria - forces the bacteria to use the best energy source first Other types of Regulations • Constitutive systems: - no regulation - always present - Enzymes that are needed during all types of growth - e.g. those involved in glycolysis • mRNA: - Unstable - half-life ~ 2 min sub-units - new mRNA • polycistronic mRNA • monocistronic mRNA - one operator for several genes - one operator per gene (in eukaryotes)