Complete Dominance Incomplete Dominance Codominance
advertisement

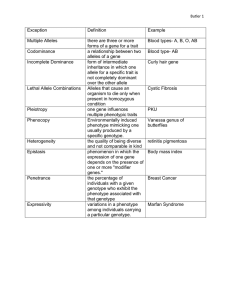
Extensions to Mendel’s Observation Types of Dominance Relationships Between Alleles of Same Locus: • Complete Dominance • Incomplete Dominance • Codominance Dominance is not always complete • Crosses between true-breeding strains can produce hybrids with phenotypes different from both parents – Incomplete dominance • F1 hybrids that differ from both parents express an intermediate phenotype. Neither allele is dominant or recessive to the other • Phenotypic ratios are same as genotypic ratios – Codominance • F1hybrids express phenotype of both parents equally • Phenotypic ratios are same as genotypic ratios Summary of dominance relationships Fig. 3.2 Complete Dominance: • Difficult to explain at the molecular level. • Recessive allele may be inactive (no product) Incomplete Dominance: • Hybrid phenotype is midway between the two parents •In snapdragon, red allele produces pigment white allele produces no pigment Hybrid (red/white) produces half the amount of pigment, hence pink in color Codominance: Each allele in the hybrid expresses itself clearly • Coat color in lentils • Blood groups A and B. Multiple Alleles: A gene locus may have more than two alleles but only two can be present in a diploid individual. Examples: • ABO blood groups: one gene locus, 3 alleles, IA = IB > i • Seed coat patterns in lentils: one gene locus, 5 alleles, marbled-1 > marbled-2 > spotted = dotted > clear • Histocompatibility antigens: 3 gene loci HLA-A, HLA-B and HLA-C. Each gene locus has 20-100 alleles. Every pair of alleles within a locus show co-dominance creating an astronomic number of variations How do new alleles in any one gene arise? By mutations. When a mutation causes one specific change in a gene (such as a single nucleotide substitution at the DNA level) this produces a new form of the gene i.e. a new allele. Distribution of alleles in nature: A wild type allele for any gene locus has a frequency of >1%. Based on that, two types of genes exist in natural populations: 1. Monomorphic: have only one wild type allele. 2. Polymorphic: have more than one wild type allele. Monomorphic: The agouti gene in mice is one of the main genes determining coat color in mice. Three alleles are known for this gene A gives rise to agouti color (each hair striped with yellow & black) at gives rise to black belly/yellow body fur a gives rise to all black fur Dominance series: A > at > a A is the wild type allele and its frequency in nature is >99%. This is because at and a make the mice easily seen by predators so they die before they reach the reproductive age. Polymorphic: 1. ABO blood groups: IA, IB and i each has a frequency of >1%. So they are all wild type alleles. 2. Self incompatibility gene in tomato and petunia has a series of alleles. This series promotes out-crossing and encourages the propagation of new mutant alleles in this gene locus. So there are several wild type and mutant alleles for this gene Pleiotropy: One gene contributes to several visible characteristics that are not related. 1. Example is a single recessive allele causes respiratory problems and sterility in men of a tribe in New Zealand. How? This recessive allele encodes for a defective protein required for the motion of cilia in the respiratory tract and also for flagella movement in sperms. 2. A recessive lethal allele in mice AY also produces a dominant yellow coat color phenotype In humans, Tay-Sachs disease is caused by a recessive lethal that encodes a defective hexoseaminidase, an important enzyme needed to rid the body from neurotoxic metabolites. Heterozygotes (carriers) can be easily detected by assaying the levels of hexoseaminidase activity in the blood. They have half the amount in a homozygous normal person. Gene Interactions: Two genes can interact to produce one trait. 1. New phenotypes are produced by the combined action of the alleles of two different gene loci. a) seed coat color in lentils (9:3:3:1) b) flower color in sweet peas result from complementary genes (9:7) 2. Alleles of one gene locus mask the effects of alleles from a different gene locus (Epistasis) a) Recessive epistasis (9:3:4) b) Dominant epistasis (12:3:1 or 13:3) Heterogenous Traits: • homozygosity of mutations at many gene loci can cause the trait (e.g. deafness in humans) • complementation test is useful in heterogenous traits because it can reveal whether two defective individuals (for the trait) have mutations in the same gene locus or in different loci Quantitative traits: • Unlike a Mendelian trait which is controlled by a single gene locus (monogenic), a quantitative trait is controlled by 3 or more gene loci (polygenic). • The contribution of each gene to the quantitative trait is additive leading to the appearance of many phenotypic classes in F2. With many genes it becomes difficult to separate the phenotypic classes in F2 and the trait is known as continuous. Example is human height and crop yield.