The General Linear Model and Statistical Parametric Mapping
advertisement

Sensor & Source Space Statistics
Rik Henson
(MRC CBU, Cambridge)
With thanks to Jason Taylor, Vladimir Litvak, Guillaume Flandin,
James Kilner & Karl Friston
Overview
A mass-univariate statistical approach to localising
effects in space/time/frequency (using replications
across trials/subjects)…
Overview
• Sensor Space:
1. Random Field Theory (RFT)
2. 2D Time-Freq (within-subject)
3. 3D Scalp-Time (within-subject)
4. 3D Scalp-Time (between-subjects)
• Source Space:
1. 3D contrast images
2. SPM vs SnPM vs PPM (vs FDR)
3. Other issues & Future directions
4. Multivariate
1. Random Field Theory (RFT)
RFT is a method for correcting for multiple statistical
comparisons with N-dimensional spaces (for parametric
statistics, eg Z-, T-, F- statistics)…
1. When is there an effect in time, eg GFP (1D)?
2. Where is there an effect in time-frequency space (2D)?
3. Where is there an effect in time-sensor space (3D)?
4. Where is there an effect in time-source space (4D)?
Worsley Et Al (1996). Human Brain Mapping, 4:58-73
2. Single-subject Example
• “Multimodal” Dataset in SPM8
manual (and website)
• Single subject:
128 EEG
275 MEG
3T fMRI (with nulls)
1mm3 sMRI
• Two sessions
• ~160 face trials and ~160
scrambled trials per session
• (N=12 subjects soon, as in
Henson et al, 2009 a, b, c)
Chapter 33, SPM8 Manual
2. Where is an effect in time-frequency (2D)?
Faces
• Single MEG channel
• Mean over trials of Morlet Wavelet
projection (i.e, induced + evoked)
• Write as t x f x 1 image per trial
• SPM, correct on extent / height
Faces > Scrambled
Scrambled
Kilner Et Al (2005) Neurosci. Letters
Chapter 33, SPM8 Manual
3. Where is an effect in scalp-time space (3D)?
• 2D sensor positions specified or
projected from 3D digitised positions
• Each sample projected to a 32x32 grid
using linear interpolation
• Samples tiled to created a 3D volume
Chapter 33, SPM8 Manual
t
y
x
• F-test of means of ~150 EEG trials of
each type (since polarity not of interest)
• (Note that clusters depend on reference)
3. Where is an effect in scalp-time space (3D)?
More sophisticated 1st-level design matrices, e.g, to remove trial-by-trial confounds within
each subject, and create mean adjusted ERP for 2nd–level analysis across subjects
Each trial-type (6)
Confounds (4)
Across-subjects
(2nd-level)
Each trial
Within-subject
(1st-level)
beta_00* images reflect mean (adjusted)
3D scalp-time volume for each condition
Henson Et Al (2008) Neuroimage
4. Where is an effect in scalp-time space (3D)?
Mean ERP/ERF images can also be tested between-subjects. Note however for
MEG, some alignment of sensors may be necessary (e.g, SSS, Taulu et al, 2005)
Without transformation to Device Space
Stats over 18 subjects on RMS of 102 planar gradiometers
With transformation to Device Space
Taylor & Henson (2008) Biomag
Overview
• Sensor Space:
1. Random Field Theory (RFT)
2. 2D Time-Freq (within-subject)
3. 3D Scalp-Time (within-subject)
4. 3D Scalp-Time (between-subjects)
• Source Space:
1. 3D contrast images
2. SPM vs SnPM vs PPM (vs FDR)
3. Other issues & Future directions
4. Multivariate
Where is an effect in source space (3D)?
Source analysis of N=12 subjects; 102 magnetometers; MSP; evoked; RMS; smooth 12mm
1. Estimate evoked/induced energy
(RMS) at each dipole for a certain
time-frequency contrast (e.g, from
sensor stats, e.g 0-20Hz, 150200ms), for each condition (e.g,
faces & scrambled) and subject
Analysis
Mask
2. Smooth along the 2D surface
3. Write these data into a 3D image
in MNI space (if canonical /
template mesh used)
4. Smooth by 8-12mm in 3D (to
allow for normalisation errors)
Henson Et Al (2007) Neuroimage
Note sparseness of
MSP inversions….
Where is an effect in source space (3D)?
Source analysis of N=12 subjects; 102 magnetometers; MSP; evoked; RMS; smooth 12mm
1. Classical SPM approach
Caveats:
• Inverse operator induces
long-range error correlations
(e.g, similar gain vectors from
non-adjacent dipoles with
similar orientation), making
RFT conservative
• Need a cortical mask, else
activity “smoothed” outside
• Distributions over subjects
may not be Gaussian…
SPM
p<.05 FWE
Where is an effect in source space (3D)?
Source analysis of N=12 subjects; 102 magnetometers; MSP; evoked; RMS; smooth 12mm
2. Nonparametric, SnPM
• Robust to non-Gaussian
distributions
• Less conservative than RFT
when dfs<20
Caveats:
• No idea of effect size (e.g, for
future experiments)
• Exchangeability difficult for
more complex designs
SnPM
p<.05 FWE
Where is an effect in source space (3D)?
Source analysis of N=12 subjects; 102 magnetometers; MSP; evoked; RMS; smooth 12mm
3. PPMs
PPM
p>.95 (γ>1SD)
• No need for RFT (no MCP!)
• Threshold on posterior
probability of an effect
(greater than some size)
• Can show effect size after
thresholding…
Caveats:
• Assume normal distributions
(e.g, of mean over voxels);
sometimes not met for MSP
(though usually fine for IID)
Grayscale=
Effect Size
Where is an effect in source space (3D)?
Source analysis of N=12 subjects; 102 magnetometers; MSP; evoked; RMS; smooth 12mm
4. FDR?
• Topological issues…?
SPM
p<.05 FWE
Where is an effect in source space (3D)?
Some further thoughts:
• Since data live in sensor space, why not perform stats there, and just report
some mean localisation (e.g, across subjects)?
True but:
What if sensor data not aligned (e.g, MEG)? (Taylor & Henson, 2008)?
What if want to fuse modalities (e.g, MEG+EEG) (Henson et al, 2009)?
What if want to use source priors (e.g, fMRI) (Henson et al, submitted)?
• Contrast localisations of conditions, or localise contrast of conditions?
“DoL” or “LoD” (Henson et al, 2007, Neuroimage)
LoD has higher SNR (though difference only lives in trial-average, i.e evoked)?
But how test localised energy of a difference (versus baseline?)
Construct inverse operator (MAP) from a difference, but then apply that operator
to individual conditions (Taylor & Henson, in prep)
Future Directions
• Extend RFT to 2D cortical surfaces (“surfstat”)
Pantazis Et Al (2005) NeuroImage
• Go multivariate…
–
To localise (linear combinations) of spatial (sensor or source)
effects in time, using Hotelling-T2 and RFT
Carbonell Et Al (2004) NeuroImage
–
To detect spatiotemporal patterns in 3D images (MLM / PLS)
Duzel Et Al (2003) Neuroimage
Kherif Et Al (2004) NeuroImage
Multivariate Model (MM) toolbox
Famous
Novel
Scrambled
Multivariate Linear Model
(MLM) across subjects on
MEG Scalp-Time volumes
(now with 3 conditions)
Famous
Novel
Scrambled
Sensitive (and suggestive
of spatiotemporal dynamic
networks), but “imprecise”
X
“M170”?
Kherif Et Al (2004) NeuroImage
The End
2. Where is an effect in time-frequency (2D)?
Kilner Et Al (2005) Neurosci. Letters
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Weighted Minimum Norm, Regularisation
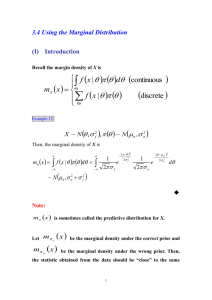
Linear system to be inverted:
Y LJ E
E ~ N (0, Ce )
Y = Data, n sensors x t=1 time-samples
J = Sources, p sources x t time-samples
L = Forward model, n sensors x p sources
E = Multivariate Gaussian noise, n x t
Ce= error covariance over sensors
Since n<p, need to regularise, eg “weighted minimum (L2) norm” (WMN):
J arg min {|| Ce1/ 2 (Y LJ ) ||2 || WJ ||2 }
J
W = Weighting matrix
(W T W ) 1 LT [ L(W TW ) 1 K T Ce ]1Y
(Tikhonov)
||Y – LJ||2
“L-curve” method
= regularisation
(hyperparameter)
||WJ||2
W =I
W = DDT
W = diag(LTL)-1
Wp = (LpTCy-1Lp)-1
W =…
minimum norm
coherent
depth-weighted
SAM
….
Phillips Et Al (2002)
Neuroimage, 17, 287–301
Equivalent Bayesian Formulation
Equivalent “Parametric Empirical Bayes” formulation:
Y LJ E
(e )
J 0 E ( j)
(e)
(e)
E ~ N (0, C )
E ( j ) ~ N (0, C ( j ) )
Posterior is product of likelihood and prior:
p(J | Y) p(Y | J ) p( J )
Maximal A Posteriori (MAP) estimate is:
Jˆ C ( j ) LT ( LC ( j ) LT C ( e ) ) 1Y
(Contrasting with Tikhonov):
(W T W ) 1 LT [ L(W T W ) 1 LT C (e) ]1 Y
C ( j ) (W T W ) 1
Y = Data, n sensors x t=1 time-samples
J = Sources, p sources x t time-samples
L = Forward model, n sensors x p sources
C(e) = covariance over sensors
C(j) = covariance over sources
Y (1) X (1) ( 2 ) E (1)
( 2 ) X ( 2 ) ( 3) E ( 2 )
( 3) 0...
W = Weighting matrix
W =I
W = DDT
W = diag(LTL)-1
Wp = (LpTCy-1Lp)-1
W =…
minimum norm
coherent
depth-weighted
SAM
….
Phillips Et Al (2005)
Neuroimage, 997-1011
Covariance Constraints (Priors)
How parameterise C(e) and C(j)?
“IID” constraint on sensors (Q(e)=I(n))
C ( e) (i e )Qi( e )
i
C ( j ) (i j )Qi( j )
Q = (co)variance components (Priors)
λ = estimated hyperparameters
# sensors
# sensors
i
Sparse priors on sources (Q1(j), Q2(j), …)
“IID” constraint on sources (Q(j)=I(p))
# sources
…
# sources
# sources
# sources
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Expectation-Maximisation (EM)
How estimate λ? …. Use EM algorithm:
(i )
T ~
ˆ
j EM YY , Q
~
Q Q1( e ) , Q2( e ) ,..., LQ1( j ) LT , LQ2( j ) LT ,...
…to maximise the (negative) “free energy” (F):
F 12 tr(C 1YY T ) 2t ln | C | pt2 ln | 2 |
C LC ( j ) LT C ( e )
C (i ) (ji )Q(ji )
(Note estimation in nxn sensor space)
Once estimated hyperparameters (iterated M-steps), get MAP for parameters (single E-step):
Jˆ MY
M Cˆ ( j ) LT Cˆ 1
(Can also estimate conditional covariance of parameters, allowing inference:)
ˆ Cˆ ( j ) MLCˆ ( j )
Phillips et al (2005) Neuroimage
Multiple Constraints (Priors)
Multiple constraints: Smooth sources (Qs), plus valid (Qv) or invalid (Qi) focal prior
Qs
500 simulations
Qs
Qs,Qv
500 simulations
Qs,Qi
Qs,Qi,Qv
Qv
Qi
Mattout Et Al (2006)
Neuroimage, 753-767
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Model Evidence
A (generative) model, M, is defined by the set of {Q(e), Q(j), L}:
The “model log-evidence” is bounded by the free energy:
ln p(Y M ) ln p(Y , J | M )dJ F
Friston Et Al (2007)
Neuroimage, 34, 220-34
(F can also be viewed the difference of an “accuracy” term and a “complexity” term):
F accuracy complexity 12 tr(C 1YY T ) 2t ln | C |
pt
2
ln | 2 |
Two models can be compared using the “Bayes factor”:
p(Y | M 1 )
p(Y | M 2 )
Also useful when comparing different forward models, ie L’s, Henson et al (submitted-b)
Model Comparison (Bayes Factors)
Multiple constraints: Smooth sources (Qs), plus valid (Qv) or invalid (Qi) focal prior
Qs
Qs,Qv
Qs,Qv,Qi
(Qs,Qi)
LogEvidence
205.2
214.1
214.7
204.9
Mattout Et Al (2006)
Neuroimage, 753-767
Bayes
Factor
7047
Qs
Qv
1.8
(1/9899)
Qi
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Temporal Correlations
To handle temporally-extended solutions, first assume temporal-spatial factorisation:
~
Y LJ E
E ~ N (0,V C )
J ~ N (0,V ( j ) C ( j ) )
(e)
(e)
~
Y = vectorised data, nt x 1
C(e) = spatial error covariance over sensors
V(e)= temporal error covariance over sensors
C(j) = spatial error covariance over sources
V(j) = temporal error covariance over sources
In general, temporal correlation of signal (sources) and noise (sensors) will differ, but can
project onto a temporal subspace (via S) such that:
S TVe S S TV j S S TVS
V typically Gaussian autocorrelations…
V KK T
(i j ) 2
K ( ) ij exp
2
2
~ 4ms
Friston Et Al (2006) Human
Brain Mapping, 27:722–735
S typically an SVD into Nr temporal modes…
Then turns out that EM can simply operate on
prewhitened data (covariance), where Y size n x t:
ˆ EM (
1
YS ( S T VS ) 1 S T Y T , Q)
Nr
Jˆ MYSS T
Localising Power (eg induced)
Friston Et Al (2006) Human
Brain Mapping, 27:722–735
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Automatic Relevance Detection (ARD)
When have many constraints (Q’s), pairwise model comparison becomes arduous
Moreover, when Q’s are correlated, F-maximisation can be difficult (eg local maxima),
and hyperparameters can become negative (improper for covariances)
Prestim Baseline
Anti-Averaging
Smoothness
Depth-Weighting
Sensor-level
Note: Even though Qs may be
uncorrelated in source space, they
can become correlated when
projected through L to sensor space
(where F is optimised)
Henson Et Al (2007)
Neuroimage, 38, 422-38
Source-level
C LC ( j ) LT C ( e )
Automatic Relevance Detection (ARD)
When have many constraints (Q’s), pairwise model comparison becomes arduous
Moreover, when Q’s are correlated, F-maximisation can be difficult (eg local maxima),
and hyperparameters can become negative (improper for covariances)
To overcome this, one can:
1) impose positivity constraint on hyperparameters:
ln( ) exp( )
2) impose (sparse) hyperpriors on the (log-normal) hyperparameters:
p( ) ~ N ( , )
8
aI , a 32
Uninformative priors are then “turned-off” as 0 (“ARD”)
Complexity
1
1
F 12 tr (C 1YY T ) 2t ln | C | pt2 ln | 2 | ln | 1 | ( ) T 1 ( )
2
2
(…where η and Σλ are the posterior mean and covariance of hyperparameters)
Automatic Relevance Detection (ARD)
When have many constraints (Q’s), pairwise model comparison becomes arduous
Moreover, when Q’s are correlated, F-maximisation can be difficult (eg local maxima),
and hyperparameters can become negative (improper for covariances)
Anti-Averaging
Smoothness
Depth-Weighting
Henson Et Al (2007)
Neuroimage, 38, 422-38
Source-level
Sensor-level
Prestim Baseline
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Multiple Sparse Priors (MSP)
So why not use ARD to select from a large number of sparse source priors….!?
Q(2)1
…
Left patch
…
Right patch
…
Q(2)j
Q(2)N
Bilateral patches
…
Q(2)j+1
…
Q(2)j+2
Friston Et Al (2008) Neuroimage
Multiple Sparse Priors (MSP)
So why not use ARD to select from a large number of sparse source priors….!
No depth bias!
Friston Et Al (2008) Neuroimage
2. Parametric Empirical Bayes (PEB)
•
•
•
•
•
•
•
Weighted Minimum Norm & Bayesian equivalent
EM estimation of hyperparameters (regularisation)
Model evidence and Model Comparison
Spatiotemporal factorisation and Induced Power
Automatic Relevance Detection (hyperpriors)
Multiple Sparse Priors
MEG and EEG fusion (simultaneous inversion)
Fusion of MEG/EEG
Separate Error Covariance components for each of i=1..M modalities (Ci(e)):
~
~
Y1 L1
E1( e )
~ ~
(e)
Y
L
2 2 J E2
~ ~
(e)
E M
YM LM
Data and leadfields scaled
(with mi spatial modes):
Yi
Li
Yi
1
mi
T
tr (YY
i i )
Li
1
mi
tr ( Li LTi )
C (e)
C1( e )
0
0
0
C2( e )
0
0
(e)
0 Cd
Ci( e ) (jie )Qij( e )
j
Remember, EM returns conditional precisions
(Σ) of sources (J), which can be used to
compare separate vs fused inversions…
ˆ C ( j ) MLC ( j )
Henson Et Al (2009b) Neuroimage
Fusion of MEG/EEG
Magnetometers (MEG)
Gradiometers (MEG)
Electrodes (EEG)
+ Fused…
ˆ 71
ˆ 73
ˆ 98
ˆ 111
Henson Et Al (2009b) Neuroimage
Overview
1. Random Field Theory for Space-Time images
2. Empirical Bayesian approach to the Inverse Problem
3. A Canonical Cortical mesh and Group Analyses
4. [ Dynamic Causal Modelling (DCM) ]
3. Canonical Mesh & Group Analyses
•
•
•
•
A “canonical” (Inverse-normalised) cortical mesh
Group analyses in 3D
Use of fMRI spatial priors (in MNI space)
Group-based inversions
A “Canonical” Cortical Mesh
Given the difficulty in (automatically) creating accurate cortical meshes from
MRIs, how about inverse-normalising a (quality) template mesh in MNI space?
Original
MRI
Normalised
MRI
Spatial Normalisation
Template
MRI
(in “MNI”
space)
Ashburner & Friston
(2005) Neuroimage
Warps…
A “Canonical” Cortical Mesh
N=1
Apply inverse of warps from spatial normalisation
of whole MRI to a template cortical mesh…
Individual
Canonical
Template
Individual
Canonical
Template
“Canonical”
Mattout Et Al (2007) Comp.
Intelligence & Neuroscience
A “Canonical” Cortical Mesh
N=9
But warps from cortex not appropriate
to skull/scalp, so use individually (and
easily) defined skull/scalp meshes…
CanInd
Canonical Cortex
Individual Skull
Individual Scalp
Statistical tests of model evidence
over N=9 MEG subjects show:
1. MSP > MMN
2. BEMs > Spheres (for CanInd)
3. (7000 > 3000 dipoles)
4. (Normal > Free for MSP)
Free Energy/104
Henson Et Al (2009a) Neuroimage
3. Canonical Mesh & Group Analyses
•
•
•
•
A “canonical” (Inverse-normalised) cortical mesh
Group analyses in 3D
Use of fMRI spatial priors (in MNI space)
Group-based inversions
Group Analyses in 3D
Once have a 1-to-1 mapping from M/EEG source to MNI space, can create 3D
normalised images (like fMRI) and use SPM machinery to perform group-level
classical inference…
N=19, MNI space, Pseudowords>Words
300-400ms with >95% probability
Smoothed,
Interpolated
J
Taylor & Henson (2008), Biomag
3. Canonical Mesh & Group Analyses
•
•
•
•
A “canonical” (Inverse-normalised) cortical mesh
Group analyses in 3D
Use of fMRI spatial priors (in MNI space)
Group-based inversions
fMRI spatial priors
…
Group fMRI results in MNI
space can be used as
spatial priors on individual
source space...
Thresholding and connected component labelling
…
...importantly each fMRI
cluster is separate prior, so
is “weighted” independently
Project onto cortical surface using Voronoï diagram
Henson Et Al (submitted)
…
Prior covariance components
Qj
3. Canonical Mesh & Group Analyses
•
•
•
•
A “canonical” (Inverse-normalised) cortical mesh
Group analyses in 3D
Use of fMRI spatial priors (in MNI space)
Group-based inversions
Group-based source priors
Concatenate data and leadfields over i=1..N subjects…
~
Y1 L1
E1 …projecting data and leadfields to a reference subject (0):
~
E
L
Y
2 2 J 2
~
T
T 1
Y
A
L
L
(
L
L
)
i Ai Yi
i
0 i
i i
~
L
Y
N N
E N
Common source-level priors:
C
( j)
Q
( j)
k
( j)
k
Subject-specific sensor-level priors:
Ci(e) (ike) AiQk(e) AiT
C (e)
C1( e )
0
0
0
C2( e )
0
0
(e)
0 C N
C L0C ( j ) LT0 C ( e )
Litvak & Friston (2008), Neuroimage
Group-based source priors
N=19, MNI space, Pseudowords>Words, 300-400ms with >95% probability
Individual Inversions
Group Inversion
Taylor & Henson (in prep)
Summary
SPM also implements Random Field Theory for principled
correction of multiple comparisons over space/time/freq
SPM implements a variant of the L2-distributed norm that:
1.
2.
3.
4.
5.
effectively automatically “regularises” in principled fashion
allows for multiple constraints (priors), valid & invalid
allows model comparison, or automatic relevance detection…
…to the extent that multiple (100’s) of sparse priors possible
also offers a framework for MEG+EEG fusion
SPM can also inverse-normalise a template cortical mesh that:
1. obviates manual cortex meshing
2. allows use of fMRI priors in MNI space
3. allows using group constraints on individual inversions