File
advertisement

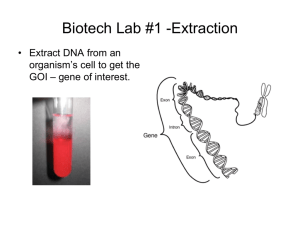
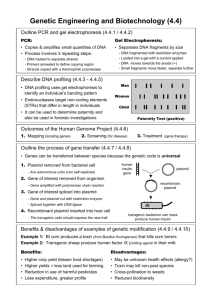
RECOMBINANT DNA TECHNIQUE Objective Students will model the process of using restriction enzymes and plasmids to form recombinant DNA. Background Information major tools of recombinant DNA technology are bacterial enzymes called restriction enzymes. Each enzyme recognizes a short, specific nucleotide sequence in DNA Result: a set of double-stranded molecules DNA fragments with singlestranded ends, called “sticky ends”. Sticky ends are not really sticky bases on the sticky ends form base pairs with the complementary bases on other DNA molecules. Enzyme cuts the backbones of the molecules at that sequence. sticky ends of DNA fragments can be used to join DNA pieces originating from different sources. In order to be useful, the recombinant DNA molecules have to be made to replicate and function genetically within a cell. Small DNA fragments can be inserted into the plasmids, which are then introduced into bacterial cells. One method for doing this is to use plasmid DNA from bacteria. As the bacteria reproduce, so do the recombinant plasmids. The result is a bacterial colony in which the foreign gene has been cloned. Key Ideas Plasmids - ideal vectors for genetic engineering replicate in the bacteria cell gene promoter antibiotic resistance as a selectable marker, can be transferred into bacteria by conjugation and transformation Restriction enzymes – key to the creation of a recombinant plasmid. cut DNA at specific sequences sticky ends allow strands to join TG B-10, C-10 Science background-Plasmids Bacteria cells produce restriction enzymes as a defense against bacteriophages BamHI: Bacillus amyloliquefaciens HindIII: Haemophilus influenza Where do we get restriction enzymes? TG B-12, C-12 Science background An enzyme that cuts DNA at a specific site (sequence) Blunt Ends “Sticky” Ends What is a restriction enzyme? Cut at Palindromes: the complementary DNA is a palindrome to the DNA strand Palindromes: Same forwards/backward: A nut for a jar of tuna Eva, can I stab bats in a cave? How do restriction enzymes cut? Sticky ends: want to form hydrogen bonds H-Bonds form easily Why are sticky ends important for ligation? Scientists can build designer plasmids that contain specific restriction sites This allows scientist to cut out and recombine genes to allow for cloning and gene expression. Why use restriction enzymes in Biotechnology? Cloning vector plasmid Engineered to replicate in high numbers within bacteria Plasmids Expression vector plasmid Carries the gene of interest in a specific location that allows the bacteria to express the gene Cloning Vector plasmid Contains: 1. Origin of replication 2. Selectable marker gene 3. Multiple cloning site Expression vector plasmid Contains 1. Origin of replication 2. Selectable marker gene 3. An inducible promoter 4. Transcription and translation initiation sequence pARA-R plasmid A sequence for initiating DNA replication Regulates expression of arabinose genes Promoter site Ampicillin resistant Restriction digest of pARA-R Biotech Experience Recombinant plasmid of interest pARA-R 5,302 bp PBAD-rfp 806 bp Restriction analysis of pARA-R Biotech Experience Restriction fragments after digest with Hind III and BamH I BamH I Hind III 4,496 bp Hind III BamH I 806 bp Clone That Gene activity RM2 and RM3 1. Cut the plasmid and the human DNA with the appropriate restriction enzyme 2. Insert the insulin gene into the plasmid DNA 3. Determine which antibiotic you would use to identify bacteria that have taken in the plasmid Plasmid DNA Human DNA Materials For each group: Handout: Plasmid Base Sequence Strips Handout: DNA Base Sequence Strips Handout: Restriction Enzyme Sequence Cards Scissors Tape Pencil Paper Instructions 1.Cut out the plasmid strips along the dotted lines. 2. Show me the strips and tape them together to form a single long strip. Reminder: The letters should all be in the same direction when the strips are taped. The two ends of the strip should then be taped together with the genetic code facing out to form a circular plasmid. 3. Cut out the DNA base sequence strips, and tape them together to form one long strip. The pieces must be taped together in the order indicated at the bottom of each strip. 4. Cut out the restriction enzyme cards. The enzyme cards illustrate a short DNA sequence that shows the that shows the sequence that each particular enzyme cuts. Instructions 5. Compare the sequence of base pairs on an enzyme card with the sequences of the plasmid base pairs. If you find the same sequence of pairs on both the enzyme card and the plasmid strip, mark the location on the plasmid with a pencil, and write the enzyme number in the marked area. Do this for each enzyme card. Some enzyme sequences may not have a corresponding sequence on the plasmid, and that some enzyme sequences may have more than one corresponding sequence on the plasmid. Instructions 6. Once you have identified all corresponding enzyme sequences on the plasmid, identify those enzymes which cut the plasmid once and only once. Discard any enzymes that cut the plasmid in the shaded plasmid replication sequence. Record your findings on a separate piece of paper. Instructions 7. Next, compare the enzymes you listed against the cell DNA strip. Look for any enzymes that will make two cuts in the DNA, one above the shaded insulin gene sequence and one below the shaded insulin gene sequence. Mark the areas on the DNA strip that each enzyme will cut. Instructions 8. After you have compared each enzyme with the DNA strip, select one enzyme to use to make the cuts. The goal is to cut the DNA strand as closely as possible to the insulin gene sequence without cutting into the gene sequence. Make cuts on both the plasmid and the DNA strips. Make the cuts in the staggered fashion indicated by the black line on the enzyme card. Tape the sticky ends (the staggered ends) of the plasmid to the sticky ends of the insulin gene to create your recombinant DNA. Discussion Questions 1. Why was it important to find an enzyme that would cut the plasmid at only one site? What could happen if the plasmid were cut at more than one site? 2. Why was it important to discard any enzymes that cut the plasmid at the replication site? 3. Why might it be important to cut the DNA strand as closely to the desired gene as possible? 4. In this activity, you incorporated an insulin gene into the plasmid. How will the new plasmid DNA be used to produce insulin? Discussion Questions 1. Why was it important to find an enzyme that would cut the plasmid at only one site? What could happen if the plasmid were cut at more than one site? Cutting at only one site is important for controlling the variables that will be reproduced. If the restriction enzyme cut more than one site, then the plasmid might recombine with different DNA fragments. Discussion Questions 2. Why was it important to discard any enzymes that cut the plasmid at the replication site? If the plasmid were cut at the replication site, it would not be able to reproduce and transfer genetic information to its host bacterial cell. Discussion Questions 3. Why might it be important to cut the DNA strand as closely to the desired gene as possible? To make sure that the desired information is transferred to the plasmid without adding extra unknown or undesirable sequences. Discussion Questions 4. In this activity, you incorporated an insulin gene into the plasmid. How will the new plasmid DNA be used to produce insulin? The new plasmid DNA will be introduced into bacterial cells, where it will reproduce, creating clones of the insulin gene.