Sequence - BIOTEC - Biotechnology Center TU Dresden

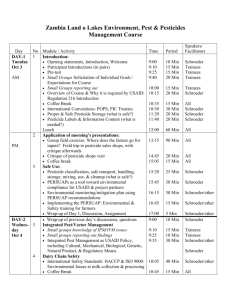
Introduction based on
Chapter 1
Lesk, Introduction to Bioinformatics
Michael Schroeder
BioTechnological Center
TU Dresden Biotec
Contents
n Molecular biology primer n The role of computer science n Phylogeny n Sequence Searching n Protein structure n Clinical implications n Read chapter 1
By Michael Schroeder, Biotec, 2
23 June 2000: Draft of Human genome sequenced!
n 1953: Watson and Crick discover the structure of DNA n 2000: Draft of human genome is published n “The most wondrous map ever produced by human kind” n “One of the most significant scientific landmarks of all time, comparable with the invention of the wheel or the splitting of the atom”
By Michael Schroeder, Biotec 3
High-throughput biomedicine
n Microarrays n Measure activity of thousands of genes at the same time n Example: n Cancer n Compare activity with and without drug treatment n Result: Hundreds of candidate drug targets n RNAi (Noble prize 2004, Fire and Mello) n Knock-down genes and observe effect n Example: n Infectious diseases n Which proteins orchestrate entry into cell?
n Result: Hundreds of candidate proteins n Atomic force microscopes (Noble prize Binnig) n Pull protein out of membrane and measure force n Example: n Eye diseases resulting fomr misfolding n Result: Hundreds of candidate residues
By Michael Schroeder, Biotec 4
Drug Discovery
n Challenge: Longer time to market, fewer drugs, exploding costs n Approach: Use of compound libraries and highthroughput screening
By Michael Schroeder, Biotec, 5
HTS and Bioinformatics
n High-throughput technologies have completely changed the work of biomedical researchers n Challenge: Interpret (often large) results of screens n Approach: Before running secondary assays use bioinformatics and IT to assemble all possible information
By Michael Schroeder, Biotec 6
>1.000.000
Sequences
By Michael Schroeder, Biotec
Good News
>30.000
3D Structures
Number of PubMed Abstracts
14,000,000
12,000,000
10,000,000
8,000,000
6,000,000
4,000,000
2,000,000
0
1960
>16.000.000
Articles
1970 1980
Year
1990 2000
Molecular Biology Database List at Nucleic Acids Research
800
700
600
500
400
300
200
100
0
>700
DBs/Tools
2000 2001 2002 2003 year
2004 2005
7
2010
Bad News: Data != Knowledge
n How to analyse data, how to integrate data?
n Comptuer science to the rescue…
By Michael Schroeder, Biotec 8
Examlpe: computer science is key for sequencing
n Human genome is a string of length 3.200.000.000
n Shotgun sequencing: Break multiple copies of string into shorter substrings n Example: n shotgunsequencing shotgunsequencing shotgunsequencing n cing en encing equ gun ing ns otgu seq sequ sh sho shot tg uenc un n Computing problem: Assemble strings
By Michael Schroeder, Biotec 9
Computer science key for sequencing
n sh n sho n shot n otgu n tg n n gun un n ns n n seq sequ n equ n n uenc encing n n en cing n ing
QUESTION: How can you handle long repetitive sequences?
Heeeeelllllllllllooooooo
QUESTION: Why was a draft announced? When was the final version ready?
By Michael Schroeder, Biotec 10
Yersinia pestis
Arabidopsis thaliana
Buchnerasp.
APS
Aquifex aeolicus
Archaeoglobus fulgidus
Borrelia burgorferi
Mycobacterium tuberculosis
Caenorhabitis elegans
Campylobacter jejuni
Chlamydia pneumoniae
Vibrio cholerae
Drosophila melanogaster
Escherichia
Thermoplasma coli acidophilum
Helicobacter pylori
Mycobacterium leprae mouse
Neisseria meningitidis
Z2491
Plasmodium falciparum
Pseudomonas aeruginosa
Ureaplasma urealyticum rat
Rickettsia prowazekii
Saccharomyces cerevisiae
Salmonella enterica
Bacillus subtilis
Thermotoga maritima
Xylella fastidiosa
By Michael Schroeder, Biotec 11
Break through of the year 2000
Next quest:
Sequencing a genome for 1000$
By Michael Schroeder, Biotec 12
Quantity and quality of data lead to ambitious goals
n Understand integrative aspects of the biology of organisms n Interrelate sequence, three-dimensional structure, interactions, function of proteins, nucleic acids and protein-nucleic acid complexes n Travel in time n backward (deduce events in evolutionary history ) and n forward ( deliberate modification of biological systems) n Applications in medicine, agriculture, and other scientific fields
By Michael Schroeder, Biotec 13
Scenario
n New virus (e.g. SARS) and goal to develop treatment n Scientists isolate genetic material of virus n Screen genome for relationships with previously studied viruses [10] n From virus’ DNA they compute the proteins it produces [1] n Compute proteins’ three-dimensional structure and thereby obtain clues about their functions n Screen for similar proteins sequences with known structure [15] n If any are found n Then interpret difference (homology modelling) [25] n Else predict structure from sequence [55] n Identify or design small molecule blocking relevant active sites of the protein [50] n Design antibodies to neutralize the virus [50] n Index of problem difficulty: n <30: solution exists already, n >30: we cannot solve this (yet)
By Michael Schroeder, Biotec 14
Life in Time and Space
n Life n A biological organism is a naturally-occurring, self-reproducing device that effects controlled manipulations of matter, energy and information n Time n Species evolve through n natural mutation, n recombination of genes in sexual reproduction, or n direct gene transfer n Read the past in contemporary genomes n Space n Species occupy local ecosystems n Species are composed of organisms n Organisms are composed of cells n Cells are composed of molecules
By Michael Schroeder, Biotec 15
DNA – the molecule of life
By Michael Schroeder, Biotec,
Proteins
n 20 naturally occurring amino acids in proteins n Non-polar n G glycine, A alanine, P proline, V valine n I isoleucine, L leucine, F phenylalanine, M methionine n Polar n S serine, C cysteine, T threonine, N asparagine n Q glutamine, H histidine, Y tyrosine, W tryptophan n Charged n D aspartic acid, E glutamic acid, K lysine, R arginine n Other classification n H,F,Y,W are aromatic and play role in membrane proteins n Distinguish n atg = adenine-thymine-guanine and n ATG = Alanine-Threonine-Glycine
By Michael Schroeder, Biotec, 17
The genetic code
First
Position
Second
Position
(5Õ end) T C A G
T
TTT Phe TCT Ser
TTC Phe TCC Ser
TAT Tyr TGT Cys
TAC Tyr TGC Cys
C
TTA
TTG
Leu
Leu
TCA
TCG
Ser
Ser
TAA Stop TGA Stop
TAG Stop TGG Trp
CTT Leu CCT Pro CAT His
CTC Leu CCC Pro CAC His
CGT
CGC
Arg
Arg
CTA Leu CCA Pro CAA Gln CGA Arg
CTG Leu CCG Pro CAG Gln CGG Arg
ATT Ile ACT Thr AAT Asn AGT Ser
A
G
ATC Ile
ATA Ile
ACC Thr AAC Asn AGC Ser
ACA Thr AAA Lys AGA Arg
ATG Met* ACG Thr AAG Lys AGG Arg
GTT Val GCC Ala GAT Asp GGT Gly
GTC Val GCC Ala GAC Asp GGC Gly
GTA Val GCA Ala GAA Glu GGA Gly
GTG Val GCG Ala GAG Glu GGG Gly
Third
Position
(3Õ end)
T
C
A
G
T
C
A
G
T
C
A
G
T
C
A
G
By Michael Schroeder, Biotec, 18
Protein Structure
n DNA: n Nucleotides are very similar and hence the structure of
DNA is very uniform n Proteins: n Great variety in threedimensional conformation to support diverse structure and functions n If heated, protein “unfolds” to biologically-inactive structure; in normal conditions protein folds
By Michael Schroeder, Biotec 19
Paradox
n Translation from DNA sequence to amino acid sequence n is very simple to describe , n but requires immensely complicated machinery
(ribosome, tRNA) n The folding of the protein sequence into its threedimensional structure n is very difficult to describe n But occurs spontaneously
By Michael Schroeder, Biotec 20
Central Dogma
n
DNA sequence determines protein sequence
n
Protein sequence determines protein structure
n
Protein structure determines protein function
By Michael Schroeder, Biotec 21
Observables and Data Archives
n Databases in molecular biology cover n Nucleic acid and protein sequences, n Macromolecular structures and functions n Archival databanks of biological information n DNA and protein sequences including annotations n Nucleic acid and protein structures including annotations n Protein expression patterns n Derived Databases n Sequence motifs (“signatures” of protein families) n Mutations and variants in DNA and protein sequences n Classification or relationships (e.g. hierarchy of structures) n Bibliographic databases (PubMed with 17M abstracts) n Collections n of links to web sites n of databases
By Michael Schroeder, Biotec 22
What is Bioinformatics
n Bioinformatics is the marriage of biology and information technology n Bioinformatics is an integrated multidisciplinary field n Covers computational tools and methods for managing, analysing and manipulating sets of biological data n Disciplines include: n biochemistry, genetics, structural biology, artificial intelligence, machine learning, software engineering, statistics, database theory, information visualisation, algorithm design
By Michael Schroeder, Biotec, 23
Bioinformatics
n Has three components n Creation of databases n Development of algorithms to analyse data n Use of these tools for analysing biological data
By Michael Schroeder, Biotec, 24
Databases: Types of Queries 1/2
n 1. Given a sequence (fragment), find sequences in the database that are similar to it n 2. Given a protein structure (or fragment), find protein structures in the database that are similar to it n 3. Given sequence of a protein of unknown structure, find structures in the database that adopt similar threedimensional structures n 4. Given a protein structure , find sequences in the database that correspond to similar structures.
By Michael Schroeder, Biotec, 25
Databases: Given sequence, find structure
n 3. Given sequence of a protein of unknown structure, find structures in the database that adopt similar three-dimensional structures.
But How?
n Easy: Find similar sequences with known structure!
n But: There might be similar structures, whose sequence is not similar!
n 4. Given a protein structure , find sequences in the database that correspond to similar structures.
But How?
n Easy: Find similar structures and hence sequences n But: There are so many more sequences with unknown structure that the above method will have only very limited success n 1 and 2 are solved, 3 and 4 are active fields of research
By Michael Schroeder, Biotec, 26
Databases: Types of Queries 2/2
n E.g. for which proteins of known structure involved in disease of disrupted purine biosynthesis in humans, are there related proteins in yeast?
n Solution: Virtual databases that provide transparent access to a number of underlying data sources and query and analysis tools
By Michael Schroeder, Biotec, 27
Databases: Curation and Quality
n Problems: n Given that there are primary and secondary databases, n how to control updates , n how to propagate change , n how to maintain consistency ?
n Contents (experimental results, annotations, supplementary information) all have there own source of error n Older data were limited by older techniques
By Michael Schroeder, Biotec, 28
Databases: Annotation
n Experimental data (e.g. raw DNA sequence) needs to be enriched with annotations n Source of data n Investigators responsible n Relevant publication n Feature tables (e.g. coding regions) n Problems: n (often) lack of controlled and coherent vocabulary n Computer parseable n Automated annotation needed n SwissProt = ca. 540.000 annotated sequences n TrEMBL = ca. 40 Mio unannotated sequences n Maintanence of annotations (what if error detected?)
By Michael Schroeder, Biotec, 29
Computers and Computer Science
n Relevant areas: n Artificial Intelligence n Machine Learning n Neural networks, rulebased learning n Datamining n Association rules n Software Engineering n Design, implementation, testing of software n Programming n Object-oriented C++,
Java n Imperative: C, Modula,
Pascal, Cobol, Fortran n Logic: Prolog n Funtional: ML n Scripting: Perl, Python n Statistics n Database theory n Design and maintenance of databases n How to index sequences, time series, 3D strucutres n Information Visualisation n Graph drawing, diagrams, cartoons, 3D graphics n Algorithm design n Complexity of algorithms n Efficient data structures
By Michael Schroeder, Biotec, 30
Programming
n We will use Python n Scripting language n Supports string processing well n Widely used in bioinformatics
By Michael Schroeder, Biotec, 31
Biological Classification and
Nomenclature
n Back in 18 th century, Linnaeus, a Swedish naturalist, classified living things according to a hierarchy:
Kingdom, Phylum, Class, Order, Family, Genus,
Species n Generally only genus and species are used for identification n Homo sapiens n Drosophila melanogastor n Bos taurus n Linnaeus’ classification based on observed similarity n Widely reflects biological ancestry
By Michael Schroeder, Biotec, 32
Classification of Humans and Fruit Flies
n Kingdom: n Phylum: n Class: n Order: n Family: n Genus: n Species:
Animalia
Chordata
Mammalia
Primata
Hominidae
Homo sapiens
Animalia
Chordata
Insecta
Diptera
Drosophilidae
Drosophila melanogastor
By Michael Schroeder, Biotec, 33
Homology = derived from common ancestor
n Characteristics derived from a common ancestor are called homologous n E.g. eagle’s wing and human’s arm n Other apparently similar characteristics may have arisen independently by convergent evolution n E.g. eagle’s wing and bee’s wing. The most common ancestor of eagles and bees did not have wings n Homologous characters may diverge functionally n E.g. bones in human middle and jaws of primitive fish
By Michael Schroeder, Biotec, 34
Sequence analysis and Homology
n Sequence analysis gives unambiguous evidence for relationship of species n For higher organisms sequence analysis and the classical tools of comparative anatomy, palaeontology, and embryology are often consistent n For microorganisms there are problems n Classical methods: how to describe features n Sequence analysis: lateral gene transfer
By Michael Schroeder, Biotec, 35
Domains of Life
n Ribosomal RNA is present in all organisms n Based on 15S ribosomal RNAs life is divided n Bacteria n No nucleus (procaryote) n E.g. tuberculosis and E. coli n Archaea n No nucleus (procaryote) n few organisms living in hostile environments (termophiles, halophiles, sulphur reducers, methanogens) n Eukarya n Has a nucleus contained in membrane n Nucleus contains chromosomes n Internal compartments called organelles for specialised biological processes n Area outside nucleus and organelles called cytoplasm n E.g. yeast and human beings
By Michael Schroeder, Biotec, 36
Eukaryotic cell
By Michael Schroeder, Biotec, 37
Domains of Life
By Michael Schroeder, Biotec, 38
Example: Use of sequences to determine phylogenetic relationships
Use ExPASy (www.expasy.ch) to search for pancreatic ribonuclease for horse (Equus caballus), minke whale (Balaenoptera acutorostrata), red kangaroo ( Macropus rufus )
>sp|P00674|RNP_HORSE Ribonuclease pancreatic
(EC 3.1.27.5) (RNase 1) (RNase A) - Equus caballus (Horse).
KESPAMKFERQHMDSGSTSSSNPTYCNQMMKRRNMTQGWCKPVNTF
VHEPLADVQAICLQKNITCKNGQSNCYQSSSSMHITDCRLTSGSKY
PNCAYQTSQKERHIIVACEGNPYVPVHFDASVEVST
Use sequence alignment to determine evolutionary relationship
By Michael Schroeder, Biotec, 39
Sequence alignment
1.
Global match : align all of one with all of the other sequence (mismatches, insertions, deletions)
And.
--so ,.from.hour.to.hour.we.r
ipe .and.r
ipe
|||| |||||||||||||||||||||||| ||||||
And.
then ,.from.hour.to.hour.we.r
ot.and.r
ot-
2.
Local match : find region in one sequence that matches the other (mismatches, insertions, deletions ; ends can be ignored)
My .care.is.
loss .of.care,.by.
old .care.
d on e,
||||||||| ||||||||||||| |||||| ||
Your .care.is.
gain .of.care,.by.
new .care.
w on
By Michael Schroeder, Biotec, 40
Sequence alignment
3. Motif search : find matches of short sequence in long sequence
Option: perfect,
1 mismatch, mismatches+gaps+insertions+deletions m atch
|||| for the w atch to babble and to talk is most tolerable
By Michael Schroeder, Biotec, 41
Sequence alignment
4. Multiple sequence alignment
No.sooner.---met.--------.but.they.look’d
No.sooner.look’d.--------.but.they.lo-v’d
No.sooner.lo-v’d.--------.but.they.sigh’d
No.sooner.sigh’d.--------.but.they.--asked.one.another.the.reason
No.sooner.knew.the.reason.but.they.-------------sought.the.remedy
No.sooner. .but.they.
By Michael Schroeder, Biotec, 42
Example: Multiple alignment
Use sequence alignment to determine evolutionary relationship…
Example: horse, whale and kangaroo
Expected: horse and whale are placental mammals, kangaroo is marsupial
Multiple alignment with CLUSTAL-W
(http://www.genome.jp/tools/clustalw) multiple sequence alignment computer program main parameters: gap opening/extension penalty
By Michael Schroeder, Biotec, 43
FASTA format
>sp|P00674|RNP_HORSE Ribonuclease pancreatic (EC 3.1.27.5)
(RNase 1) (RNase A) - Equus caballus (Horse).
KESPAMKFERQHMDSGSTSSSNPTYCNQMMKRRNMTQGWCKPVNTFVHEPLADVQAICLQ
KNITCKNGQSNCYQSSSSMHITDCRLTSGSKYPNCAYQTSQKERHIIVACEGNPYVPVHF
DASVEVST
>sp|P00673|RNP_BALAC Ribonuclease pancreatic (EC 3.1.27.5)
(RNase 1) (RNase A) - Balaenoptera acutorostrata (Minke whale) (Lesser rorqual).
RESPAMKFQRQHMDSGNSPGNNPNYCNQMMMRRKMTQGRCKPVNTFVHESLEDVKAVCSQ
KNVLCKNGRTNCYESNSTMHITDCRQTGSSKYPNCAYKTSQKEKHIIVACEGNPYVPVHF
DNSV
>sp|P00686|RNP_MACRU Ribonuclease pancreatic (EC 3.1.27.5)
(RNase 1) (RNase A) - Macropus rufus (Red kangaroo)
(Megaleia rufa).
ETPAEKFQRQHMDTEHSTASSSNYCNLMMKARDMTSGRCKPLNTFIHEPKSVVDAVCHQE
NVTCKNGRTNCYKSNSRLSITNCRQTGASKYPNCQYETSNLNKQIIVACEGQYVPVHFDA
YV
By Michael Schroeder, Biotec, 44
Multiple Alignment with ClustalW
(http://www.genome.jp/tools/clustalw)
CLUSTAL W (1.82) multiple sequence alignmen sp|P00674|RNP_HORSE sp|P00673|RNP_BALAC sp|P00686|RNP_MACRU
KESPAMKFERQHMDSGSTSSSNPTYCNQMMKRRNMTQGWCKPVNTFVHEPLADVQAICLQ 60
RESPAMKFQRQHMDSGNSPGNNPNYCNQMMMRRKMTQGRCKPVNTFVHESLEDVKAVCSQ 60
-ETPAEKFQRQHMDTEHSTASSSNYCNLMMKARDMTSGRCKPLNTFIHEPKSVVDAVCHQ 59
*:** **:*****: :......*** ** *.**.* ***:***:**. *.*:* *
KNITCKNGQSNCYQSSSSMHITDCRLTSGSKYPNCAYQTSQKERHIIVACEGNPYVPVHF 120
KNVLCKNGRTNCYESNSTMHITDCRQTGSSKYPNCAYKTSQKEKHIIVACEGNPYVPVHF 120
ENVTCKNGRTNCYKSNSRLSITNCRQTGASKYPNCQYETSNLNKQIIVACEG-QYVPVHF 118
:*: ****::***:*.* : **:** *..****** *:**: :::******* ******
DASVEVST 128
DNSV---- 124
DAYV---- 122
* *
By Michael Schroeder, Biotec, 45
Example: Number of Aligned Residues
Horse and Minke whale: 95
Minke whale and Red kangoroo: 82
Horse and Red kangoroo: 75
Conclusion: Horse and whale share the most identical residues
By Michael Schroeder, Biotec, 46
New Example:
Elephant and Mammoth
Mitochondrial cytochrome b from
Siberian woolly mammoth (Mammuthus primigenius) preserved in arctic permafrost
African elephant (Loxodonta africana)
Indian elephant (Elephans maximus)
Q: To which one is the Mammuth more closely related?
By Michael Schroeder, Biotec, 47
Indian elephant: sp|P24958|CYB_LOXAF
Mammoth: sp|P92658|CYB_MAMPR
African elephant: sp|O47885|CYB_ELEMA
MTHIRKSHPLLKIINKSFIDLPTPSNISTWWNFGSLLGACLITQILTGLFLAMHYTPDTM 60
MTHIRKSHPLLKILNKSFIDLPTPSNISTWWNFGSLLGACLITQILTGLFLAMHYTPDTM 60
MTHTRKFHPLFKIINKSFIDLPTPSNISTWWNFGSLLGACLITQILTGLFLAMHYTPDTM 60
*** ** ***:**:**********************************************
TAFSSMSHICRDVNYGWIIRQLHSNGASIFFLCLYTHIGRNIYYGSYLYSETWNTGIMLL 120
TAFSSMSHICRDVNYGWIIRQLHSNGASIFFLCLYTHIGRNIYYGSYLYSETWNTGIMLL 120
TAFSSMSHICRDVNYGWIIRQLHSNGASIFFLCLYTHIGRNIYYGSYLYSETWNTGIMLL 120
************************************************************
LITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTNLVEWIWGGFSVDKATLNRFFA 180
LITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTDLVEWIWGGFSVDKATLNRFFA 180
LITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTNLVEWIWGGFSVDKATLNRFFA 180
**************************************:*********************
LHFILPFTMIALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLGLLILILLLL 240
LHFILPFTMIALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLGLLILILFLL 240
FHFILPFTMVALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLGLLILILLLL 240
:********:***********************************************:**
LLALLSPDMLGDPDNYMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALLLSILI 300
LLALLSPDMLGDPDNYMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALLLSILI 300
LLALLSPDMLGDPDNYMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALFLSILI 300
******************************************************:*****
LGLMPLLHTSKHRSMMLRPLSQVLFWTLTMDLLTLTWIGSQPVEYPYIIIGQMASILYFS 360
LGIMPLLHTSKHRSMMLRPLSQVLFWTLATDLLMLTWIGSQPVEYPYIIIGQMASILYFS 360
LGLMPLLHTSKHRSMMLRPLSQVLFWTLTMDLLTLTWIGSQPVEHPYIIIGQMASILYFS 360
**:*************************: *** **********:***************
IILAFLPIAGVIENYLIK 378
IILAFLPIAGMIENYLIK 378
IILAFLPIAGMIENYLIK 378
**********:*******
By Michael Schroeder, Biotec, 48
Example: Elephant and Mammoth
Mammoth and African elephant have 10 mismatches,
Mammoth and Indian elephant 14.
Significant?
Q1: can we tell from these sequences alone that they are closely related?
Q2: differences are small – do they come from selection, random noise or drift
Strategies needed difference judging of similiarities
By Michael Schroeder, Biotec, 49
Excursion:
Similarity and Homology
Important difference:
Similarity is the measurement of resemblance of sequences
Homology: common ancestor
Similarity is gradual, homology is either true or false
Similarity = now, homology = past events
Homology is only very rarely directly observed (e.g. lab population, clinical study of viral infection)
Homology is inferred from sequence similarity
By Michael Schroeder, Biotec, 50
Example: Homology/Similarity
The assertion that the cytochrome b sequences are
homologues means that there is a common ancestor
BUT:
1. Maybe cytochrome b functionally requires so many conserved residues and will hence occur in many species ( In fact, This is not the case here)
2. Maybe cytochrome b has to function this way in elephant-like species, but in fact started out from different ancestors (i.e. convergent evolution )Mammoth are homolgues – are also ribonuclease sequences homologues? Difference is much bigger
3. Maybe mammoth and african elephant have only fewer mismatches, because Indian elephant’s DNA mutated faster
4. Maybe all of them acquired cytochrome b through a virus
( horizontal gene transfer )
By Michael Schroeder, Biotec, 51
Examples: Conclusion
Classical methods confirm that for pancreatic ribonuclease (Horse – whale - kangoroo) inferring homology from similarity is justified
But to answer whether Mammoth are closer to African or
Indian elephants is too close to call (non-significant)
Problems with inferring phylogeny from gene and protein sequence comparison
Wide range of variation (possibly below statistical significance)
Different rates of evolution for different branches of the evolutionary tree
Even if relationship - which sequence came first?
By Michael Schroeder, Biotec, 52
Inferring Phylogenies with SINES and LINES
Pylogeneticist’s dream of features:
‘all-or-none’ character
Irreversible appearance
Solution:
SINES and LINES (Short and Long Interspersed
Nuclear Elements)
Repetitive, non-coding sequences in eukaryotic genomes
>30% in human genome, >50% in some plants
SINES = 70-500 base pairs long, up to 10 6 copies
LINES up to 7000 base pairs, up to 10 5 copies
They enter genome by reverse transcription of RNA
By Michael Schroeder, Biotec, 53
A practical example:
Fatherhood
The picture shows a Southern blot of DNA from different family members, probed using a mini-satellite.
You can work out which of F1 and F2 is the father of child C, by observing which bands they have in common.
(Reproduced from "Essential Medical Genetics" by M.Connor and
M.Ferguson-Smith, with permission from Blackwell Science.)
By Michael Schroeder, Biotec, 54
Why SINES are useful in phylogeny
Either present or absent
Inserted at random in non-coding portion of genome i.e. SINE has no important function so that convergent evolution can be excluded
Presence of a SINE in two species and absence in a third implies that first two species are more closely related
SINE insertion appears to be irreversible
Temporal order
Presence of a SINE in two species and absence in a third implies that ancestor of first two species is younger than ancestor of all three
By Michael Schroeder, Biotec, 55
Example revisited
Q: What is the closest land-based relative of the whales?
Classical palaeontology links Cetacea (whales, dolphins, porpoises) with Artiodactyla
(including e.g. cattle)
Belief that Cetaceans diverged before Artiodactyla split into suborder of
Suiformes (e.g. pigs),
Tylopoda (e.g. camels, llamas),
Ruminantia (e.g. deer, cattle, goats, sheep, antelopes, giraffe)
By Michael Schroeder, Biotec, 56
Example revisited
Sequence comparison results
Based on mitochondrial DNA, pancreatic ribonuclease, fibrinogen, and others
Closest relatives of whales are hippopotamuses (share 4
SINES)
These two are closest to Ruminantia
By Michael Schroeder, Biotec, 57
Searching for Similar
Sequences with PSI-Blast
False negatives :
300 out of 1000 are not found
Any search method for sequences should be
Sensitive: pick up distant relationships
Selective: reported relationships are true
Example: database with (among others) 1000 globin sequences
Globin familiy (oxygen transport) of proteins occurs in many species
Proteins have same function and structure
But there are pairs of members of the family sharing less than 10% identical residues
Sequence Database
1000 Globin
Sequences
900 Search results
True positives:
700 out of 900 are really globins
False positives :
200 out of 900 are not globins
By Michael Schroeder, Biotec, 58
Searching for Distant Relationships with PSI-BLAST
How can we find distant relationships without increasing the false negatives?
PSI-BLAST:
Position Sensitive Iterated – Basic Linear Alignment
Sequence Tool
Identifies conserved patterns within the sequences
Improves Sens and Spec
Score via intermediaries may be better than score from direct comparison
A
50%
B
50%
C
Only 10%
By Michael Schroeder, Biotec, 59
PSI-BLAST Example
Human PAX-6 gene (SwissProt ID P26367) has homologues in many different species
(human, Drosophila, etc.)
TF for eye development
Mutations in:
Human: no or deformed iris
Drosophila: no eyes, expressed in wing or leg ectopic eyes
PSI-Blast at NCBI site (www.ncbi.nlm.nih.gov)
By Michael Schroeder, Biotec, 60
Result
By Michael Schroeder, Biotec, 61
Result
• Description of sequence
• Max score – linked to data that show where sequences match
• Total score - includes scores from non-contiguous portions of the subject sequence that match the query
• Query coverage
• Identity - % of a sequence with the highest percentage of identical bases
• E-Value
• Accession number – linked to Gene bank record
By Michael Schroeder, Biotec, 62
Result
BLASTP 2.2.28+
RID: 6D2U321501N
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF excluding environmental samples from WGS projects
33,121,465 sequences; 11,555,699,950 total letters
Query= gi|6174889|sp|P26367.2|PAX6_HUMAN RecName: Full=Paired box protein
Pax-6; AltName: Full=Aniridia type II protein; AltName:
Full=Oculorhombin
Length=422
Score E
Sequences producing significant alignments: (Bits) Value ref|NP_000271.1| paired box protein Pax-6 isoform a [Homo sap... 870 0.0 ref|XP_004264012.1| PREDICTED: paired box protein Pax-6 isofo... 869 0.0 ref|XP_003910122.1| PREDICTED: paired box protein Pax-6 isofo... 869 0.0 ref|XP_004683008.1| PREDICTED: paired box protein Pax-6 isofo... 869 0.0 ref|XP_005064880.1| PREDICTED: paired box protein Pax-6 isofo... 868 0.0 ref|NP_001035735.1| paired box protein Pax-6 [Bos taurus] >re... 868 0.0 gb|AAA59962.1| oculorhombin [Homo sapiens] 868 0.0 ref|NP_037133.1| paired box protein Pax-6 [Rattus norvegicus]... 868 0.0 gb|EAW68233.1| paired box gene 6 (aniridia, keratitis), isofo... 869 0.0
...
By Michael Schroeder, Biotec, 63
Introduction to Protein Structure
Proteins play a variety of roles:
Structural (viral coat proteins, horny outer layer of human and animal skin, cytoskeleton)
Catalysis of chemical reactions (enzymes)
Transport and Storage (e.g. haemoglobin)
Regulation (e.g. hormones)
Receptor and signal transduction
Genetic transcription
Recognition (cell adhesion molecules)
Antibodies and other proteins of the immune system
By Michael Schroeder, Biotec, 64
Proteins
Are large molecules
Only small part – the active site – is functional
Evolve by structural changes produced by mutations in the amino acid sequence
Ca. 21.000 human proteins structures are now known
Overall 90.000 protein structures in PDB
Can be obtained by X-ray crystallography or nuclear magnetic resonance (NMR)
By Michael Schroeder, Biotec, 65
Structure of Proteins
Backbone and side chain
Residue i-1
, Residue i
, Residue i+1
,
S i-1
S i
S i+1
Side chain (variable)
| | |
…N-C
α
-C-N-C
α
-C-N-C
α
-C… Main chain (constant)
|| || ||
O O O
Polypeptide chain folds into a curve in space
Common structural feature
Alpha-helix
Beta-sheet
Turns and Loops
By Michael Schroeder, Biotec, 66
Hierarchy of Architecture
Primary structure : Amino acid sequence
Secondary structure : Helices, sheets, loops, hydrogen-bonding pattern of main chain
Tertiary structure : Assembly and interactions of helices, sheets, etc.
Quaternary structure : Assembly of monomers
Evolution can merge proteins
E.g.: 5 enzymes in E. coli = 1 protein in fungi Aspergillus nidulans catalyze successive steps in biosynthesis of aromatic amino acids
E.g.: Globins form tetramers in mammalian haemoglobin and dimers in ark clam Scaoharca inaequivalvis
By Michael Schroeder, Biotec, 67
Protein Structure
DHAP to GAP in Glycolyse
Triosephosphate isomerase from Bacillus stearothermophilus
Highly efficient enzyme appearing in most species
By Michael Schroeder, Biotec, 68
Extra layer of Architecture: supersecondary structure
Alpha-helix hairpin
Beta hairpin
Beta-alpha-beta unit
= Patterns of interaction between helices and sheets
By Michael Schroeder, Biotec, 69
Hierarchy of Architecture
Supersecondary structures:
Alpha-helix hairpin
Beta hairpin
Beta-alpha-beta unit
Domains:
Compact unit, single chain, independent stability
Modular proteins:
Multi-domain
Copies of related domains or “mix-and-match”
By Michael Schroeder, Biotec, 70
Classification of Protein Structure
All Alpha : mostly alpha helices
All Beta : mostly beta sheets
Alpha+Beta : Helices and sheets in different parts of the molecule, no beta-alpha-beta units
Alpha/Beta : Helices and sheets assembled from beta-alpha-beta units
Alpha/Beta linear
Alpha/Beta barrel
Little or no secondary structure
By Michael Schroeder, Biotec, 71
SCOP: Structural Classification of Proteins
top
CLASS
All alpha (284) All Beta (174) Alpha+Beta (376) Alpha/Beta (147)
FOLD
Trypsin-like serine proteases (1)
SUPERFAMILY
= evolutionary related, similar structure, not necessarily similar sequence
Immunoglobulin-like (23)
Transglutaminase (1) Immunoglobulin (6)
FAMILY
= set of domains with similar sequence
By Michael Schroeder, Biotec,
C1 set domains
(antibody constant)
V set domains
(antibody variable)
72
Pymol
By Michael Schroeder, Biotec, 73
Engrailed homeodomain (1enh)
Transcription factor important in development
Used to study protein folding
Utrophin calmodulin homology domain (1bhd)
Actin binding
Closely relatd to dystrophin, whose lack causes muscular dystrophies (weak muscles)
Cytochrome c, rice (1ccr)
Electron transport across mitochondrial membrane
By Michael Schroeder, Biotec,
DNA-binding domain of HIN recombinase (1hcr)
74
By Michael Schroeder, Biotec,
Engrailed homeodomain (1enh)
75
Fibronectin III domain (1fna)
Found on cell surface
Mannose-binding protein (1npl)
Barnase (1brn)
Cleaves RNA and is lethal if intracellular and not inhibited by barstar
By Michael Schroeder, Biotec,
TATA-box-binding protein (1cdw)
76
OB-domain from Lys-tRNA synthetase (1bbw)
Scytalone dehydratase (3std)
Alcohol dehydrogenase, NADbinding domain (1ee2)
Break down of alcohol into simpler compounds
By Michael Schroeder, Biotec,
Adenylate kinase (3adk)
Energy production
77
Chemotaxis receptor methyltransferase (1af7)
By Michael Schroeder, Biotec,
Pancreatic spasmolytic polypeptide (2psp)
Thiamine phosphate synthase (2tps)
78
Protein Structure Prediction and
Engineering
If sequence of amino acids contains enough information to specify three-dimensional structure of proteins, it should be possible to devise algorithm for prediction
Secondary structure prediction : Which segments of the sequence are helices, which strands?
Fold recognition : Given library of known structures with their sequences and a sequence with unknown structure , can we find the structure that is most similar
Homology modelling
Given two homologous sequences , one with one without structure.
If between 30 and 50% of the residues are identical, the structure can serve as a model
By Michael Schroeder, Biotec, 79
Critical Asessment of Structure
Prediction (CASP)
Chicken lysozyme KVFGRCELAAAMKRHGLDNYRGYSLGNWVCAAKFESNFNTQATNRNTDGS
Baboon alpha-lactalbumin KQFTKCELSQNLY--DIDGYGRIALPELICTMFHTSGYDTQAIVEND-ES
Chicken lysozyme TDYGILQINSRWWCNDGRTPGSRNLCNIPCSALLSSDITASVNCAKKIVS
Baboon alpha-lactalbumin TEYGLFQISNALWCKSSQSPQSRNICDITCDKFLDDDITDDIMCAKKILD
Chicken lysozyme DGN-GMNAWVAWRNRCKGTDVQA-WIRGCRL-
Baboon alpha-lactalbumin I--KGIDYWIAHKALC-TEKL-EQWL--CE-K
By Michael Schroeder, Biotec, 80
Clinical Implications of
Sequencing
Fast and reliable diagnosis of disease and risk:
Easy diagnosis (with symptoms)
In advance of appearance (e.g. Huntington)
In utero diagnosis (e.g. cystic fibrosis: thick secretions in lung)
Genetic counselling
Customized treatment (predict response to therapy/side effects)
E.g. childhood leukaemia is treated with toxic drug 6-mercaptopurine.
Small fraction of patients used to die as they lack enzyme thiopurine methyltransferase.
Identify drug targets
Nowadays targets are: ½ receptors, ¼ enzymes, ¼ hormones
7% have unknown targets
Gene therapy
Replace defective genes or supply gene products (insulin for diabetes and Blood Factor VIII for haemophilia)
However : Most diseases do not have a single genetic cause!
By Michael Schroeder, Biotec, 81
Quick check
By now you should
Have read chapter 1
Know the main data sources (sequence and structure)
Know the role that bioinformatics plays
Understand the difference between homology and similarity
Understand what sequence comparison and alignment are
Understand how they can be useful for phylogenetic studies
Understand primary, secondary, tertiary structure
Be able to assess the assumptions made and the quality of data
By Michael Schroeder, Biotec, 82