Automated Theory Formation First Steps in Bioinformatics
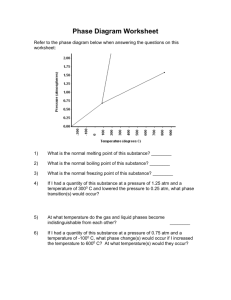
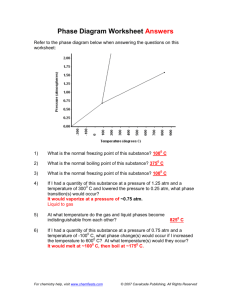
advertisement

Automated Theory Formation:
First Steps in Bioinformatics
Simon Colton
Computational Bioinformatics Laboratory
Machine Learning (ML)
Questions
Given some background information
Concepts, hypotheses (axioms)
Given some positive examples
And some negative examples
Find me an explanation
Why the positives are positive
And the negatives are negative
Example: Predictive Toxicology
Given some theory from chemistry
Structure of molecules, well known substructures
Given some examples of toxic drugs
And some examples of non-toxic drugs
Question: Why are the toxic drugs toxic?
Automated Theory Formation (ATF)
Questions
Given some background information
Concepts, hypotheses (axioms)
And some objects of interest
Numbers, Molecules, etc.
Find something interesting
Interesting things could be:
Concepts, examples, hypotheses, explanations
ATF Overview
Scientific theories contain (at least):
Concepts: salt, acid, base
Hypotheses: acid + base => salt + water
Explanations: transfer of electrons, dissolving
So, ATF should do (at least):
Concept formation, Conjecture making
Hypothesis proving and disproving.
Also needs to:
Measure interestingness, present results, etc.
HR Theory Formation System
Developed in maths
Designed to be general purpose system
Concept-based theory formation
Tries to make concept
Makes conjecture when it can’t make a concept
Tries to explain conjectures
Conjecture-based theory formation
Fix faulty conjectures with concept formation
PhD work of Alison Pease, based on Lakatos
Concept Formation in HR
10 General Production Rules
Take in old concepts, produce new concepts
Size
[a,b] : b|a
[a,n]:n = |{b:b|a}|
Split
[a] : 2|a
Negate
Split
[a]:2=|{b:b|a}|
Compose [a] : not 2|a
[a]:2=|{b:b|a}| & not 2|a
(Odd Prime Numbers)
Conjecture Making
Empirical checks are performed
After each attempt to invent a new concept
If the concept has no examples
Makes non-existence conjecture
If concept has same examples as previous
Makes an equivalence conjecture
If another concept subsumes the concept
Makes an implication conjecture
Conjecture Extraction
Suppose HR makes equivalence conjecture:
P(a) & Q(a) R(a) & S(a)
Extracts:
P(a) & Q(a) => R(a), P(a) & Q(a) => S(a)
R(a) & S(a) => P(a), R(a) & S(a) => Q(a)
Tries to Extract: P(a) => R(a), Q(a) => R(a), etc.
Prime implicates (require proving, though)
Important: gets Horn Clauses
Can be expressed in Prolog…..
Explanation Generation
In mathematical domains
HR relies on automated theorem provers
And Model generators
To find counterexamples
E.g., group theory: a*a=a a=id (prove easily)
In biological/chemistry domains
Possibly: visualisation tools, reaction pathways
Greatest Hits
Please ask me over coffee about:
Pre-processing constraint problems
Learning properties of quadratic residues
Inventing integer sequences
Puzzle generation
Adding to the TPTP library
Setting mathematical tutorial questions
…
Long term aim in Bioinformatics
Develop an ATF system similar to HOMER
But working in biological domains
Biologist provides little background info
In a format they are happy with
Program provides results
Intelligent, interesting, not too much,
And very little rubbish
Automated assistant for biology
Short term aim in Bioinformatics
HR can work with biological data
Takes input similar to Muggleton’s Progol
Use HR to solve ML problems
See how bad an idea that is
Use theory formation to improve ML
Integrate HR and Progol somehow
Naïve Approach to ML Tasks
Give HR the same input as Progol
Get it to form a theory
Look at the theory
Extract concepts which do well on the task
i.e., they look similar to target concept
Not a goal-based approach
Bad idea (slow)
Less Naïve Approach
Improve search using “forward look-ahead”
ICML Paper
This has evolved to “reactive search”
Uses HR’s own Java interpreter
HR reacts to certain events in theory formation
Scripts supplied by the user
HR also makes “near-conjectures”
Faster approach, but still fairly slow
Example – Mutagenesis42 Data
Mutagenesis similar to carcinogenisis
42 drugs supplied with atom-bond details
Atom type, number & charge, bond type (1-8)
13 are mutagenic (active), 29 are not active
Progol learned this concept (88% accurate)
active(A) :- bond(A,B,C,2), bond(A,D,B,1),atm(A,D,c,21,E)
1
c,21
2
?
?
HR’s Results
Using reactive search, four PRs, 30K steps
HR learned this concept:
active(A) :- bond(A,B,C,1), atm(B,F,21), bond(A,C,D,E)
Also 88% accurate
But, Progol’s answer “better”
Because higher information content (fewer ?s)
Biologists sometimes want more information
Is this really a simpler answer?
1
?,21
?
?
?
But…..
HR also made these equivalence conjectures
And extracted them (+100 more) for us
atm(B,X,21) atm(B,c,21)
atm(B,X,38) atm(B,n,38)
bond(A,B,C,X1) & atm(C,X2,38) bond(A,B,C,1) & atm(C,X3,38)
bond(A,X1,B,X2) & atm(B,X3,38) bond(A,B,X4,2), atm(B,X5,38)
We used these to re-write HR’s answer
By hand, but hope to automate
Giving us this answer:
1
2
c,21
?
n,38
Remember that Progol’s Answer was:
1
c,21
2
?
?
So, we filled in one of the blanks!
Are we making a meal of this?
Yes, possibly for the mutagenesis data
I was worried about the difficulty of this problem
In the last week I’ve written a
200-line Prolog program which runs quite fast
And can be distributed over multiple processors
And can be easily understood by biologists
And gets these results….
Template search – Results
Nice result one (88% accurate, lots of info)
1
c,21
2
n,38
o,40
2
o,40
Nice result two (95% accurate)
1
c,21
2
n,38
7
o,40
c,?
1
c,22
-0.132
1
7
?
c,195
c,22
h,3
0.145
Template Search - Assumptions
Connected substructures
Are interesting answers
Progol’s answers are all substructures
More specific substructures are not so bad
Biologists may even want lots of information
Don’t forget that they want to do science
Each learned concept will be true of
At least one active (positive) molecule
Template Search - Overview
User chooses template for substructures
?
?,?
?
?,?
?,?
User specifies how many ?s are allowed
E.g., 3 out of 8 in the above template
Algorithm starts with the first positive
Extracts all substructures in the template
Then takes the next positive,
for each substructure in the set
Add the LGG so that it fits both positives
Don’t go under the IC limit
Template Search – Final Part
For all the substructures
Take a disjunction
Which achieves the best accuracy
Distribution of this algorithm possible
We’re getting a big Linux farm
PPP – Processor Per Positive
finds substructures true of one positive
combine answers at the end
Conclusions & Future Work
Automated Theory Formation
May be useful to bioinformatics
Use HR’s theory to improve Progol’s results
Possibly by pre-processing Progol’s input
Or by post-processing the learned concept
Template search
Maybe a good idea? Possibly not new….
Not bad results for the Mutagenesis42 dataset