Process modelling, analysis and control: case studies for signal

Colloquium on Control in Systems Biology, University of Sheffield, 26 th March, 2007
Sensitivity Analysis and Experimental Design
- case study of an NFk B signal pathway
Hong Yue
Manchester Interdisciplinary Biocentre (MIB)
The University of Manchester h.yue@manchester.ac.uk
Outline
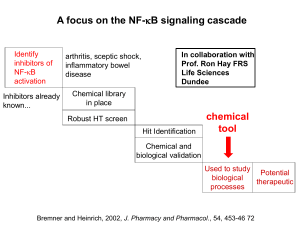
NFk B signal pathway
Time-dependant local sensitivity analysis
Global sensitivity analysis
Robust experimental design
Conclusions and future work
NFk
B signal pathway
Hoffmann et al ., Science, 298, 2002 stiff nonlinear ODE model
X
X
x
1 x
2
(state vector) x n
T
( )
0
X
0
k
1 k
2 k m
T
(parameter vector)
0.06
0.05
0.04
0.09
0.08
0.07
0.03
0.02
0.01
0
0 0.5
1
time/s
1.5
2 2.5
x 10
4
Nelson et al ., Sicence, 306, 2004
State-space model of NF-kB states definition
Characteristics of NFk
B signal pathway
Important features:
Oscillations of NFk B in the nucleus delayed negative feedback regulation by I k B a
Total NF
k B concentration x
2
3 5 7 9 x
12
x
14
x
15
x
17
x
19
x
21
0
Total IKK concentration
14
Control factors: i
8 x i
k x
61 10
Initial condition of NFk B
Initial condition of IKK
About sensitivity analysis
Determine how sensitive a system is with respect to the change of parameters
Metabolic control analysis
Identify key parameters that have more impacts on the system variables
Applications: parameter estimation, model discrimination & reduction, uncertainty analysis, experimental design
Classification: global and local dynamic and static deterministic and stochastic time domain and frequency domain
Time-dependent sensitivities (local)
Sensitivity coefficients s
x i
/
j
, s
, t
0
j
x i
0
)
Direct difference method (DDM)
S j
f X
j
f
j
( )
0
X
0
j
F j
, j
( )
0
S j
0
Scaled (relative) sensitivity coefficients s
i
j i j
x
i j
x i j
Sensitivity index
RS
1 N
N k
1 s ( )
2
Local sensitivity rankings
Sensitivities with oscillatory output
Limit cycle oscillations:
Non-convergent sensitivities
Damped oscillations: convergent sensitivities
Parameter estimation framework based on sensitivities
Dynamic sensitivities
Correlation analysis
Identifiability analysis
Robust/fragility analysis
Model reduction
Parameter estimation
Experimental design
Yue et al ., Molecular BioSystems , 2, 2006
Sensitivities and LS estimation
Assumption on measurement noise: additive, uncorrelated and normally distributed with zero mean and constant variance.
Least squares criterion for parameter estimation
J
1
2
k i
i
x k
x k
Gradient g
J
j
k i
i i
)
i
j
2
k i r i
i s i , j
( k )
Hessian matrix
2
J
j l
k i
s ( ) k
k
i r i
( k )
i
s
l
Correlation matrix
M ( ) c
Understanding correlations
K
28 and k
36 are correlated
Sensitivity coefficients for NFk
B n
.
cost functions w.r.t. ( k
28
, k
36
) and ( k
9
, k
28
).
Global sensitivity analysis: Morris method
One-factor-at-a-time (OAT) screening method
Global design : covers the entire space over which the factors may vary
Based on elementary effect (EE). Through a pre-defined sampling strategy, a number (r) of EEs are gained for each factor.
Two sensitivity measures: μ (mean), σ (standard deviation) large μ: high overall influence
(irrelevant input) large σ: input is involved with other inputs or whose effect is nonlinear
Max D. Morris, Dept. of Statistics, Iowa State University
sensitivity ranking μ-σ plane
Sensitive parameters of NFk
B model
Local sensitive Global sensitive k9, k62 k28, k29, k36, k38 k52, k61 k19, k42 k9: IKKI k
B a
-NFk
B catalytic k62: IKKI k
B a catalyst k29: I k
B a mRNA degradation k36: constituitive I k
B a translation k28: I k
B a inducible mRNA synthesis k38: I k
B a n nuclear import k52: IKKI k
B a
-NFk
B association k61: IKK signal onset slow adaptation k19: NFk
B nuclear import k42: constitutive I k
B b translation
IKK, NFk
B, I k
B a
Improved data fitting via estimation of sensitive parameters
(a) Hoffmann et al., Science (2002) (b) Jin, Yue et al., ACC2007
The fitting result of NFk
B n in the I k
B a
-NFk
B model
Optimal experimental design
Aim: maximise the identification information while minimizing the number of experiments
What to design?
Initial state values: x
0
Which states to observe: C
Input/excitation signal: u ( k )
Sampling time/rate
Basic measure of optimality:
Fisher Information Matrix
Cramer-Rao theory
T
1
FIM S Q S
ˆ i
2
FIM
1 lower bound for the variance of unbiased identifiable parameters
Optimal experimental design
Commonly used design principles:
A-optimal min trace( FIM
1
)
D-optimal
E-optimal max det( FIM ) max
min
( FIM )
Modified E-optimal design min cond( FIM )
95% confidence interval
i
1.96
i i
1.96
i
2
1
The smaller the joint confidence intervals are, the more information is contained in the measurements
Measurement set selection
Forward selection with modified E-optimal design
Estimated parameters: k , k , k , k
19 29 31 36
, k
38
, k
42
, k
52
, k
61 x
12
(IKKI k
B b
-NFk
B), x
21
(I k
B e n
-NFk
B n
), x
13
(IKKI k
B e
) , x
19
(I k
B b n
- NFk
B n
)
Step input amplitude
95% confidence intervals when :-
IKK=0.01
μM ( r ) modified E-optimal design
IKK=0.06
μM ( b ) E-optimal design
Robust experimental design
Aim: design the experiment which should valid for a range of parameter values
Measurement set selection
x x
1
, , m
1
, , m
, i m
1
i
1,
i i
FIM
i m
1
i i i
T
(nominal)
i
T
f x
i
FIM
i m
1 i
i i
T i
blkdiag (
1
, ,
m
)
(with uncertainty)
0
This gives a (convex) semi-definite programming problem for which there are many standard solvers (Flaherty, Jordan,
Arkin, 2006)
Robust experimental design
0
Uncertainty degree
(optimal design)
( middle ) (robust design)
max
(uniform design)
Conclusions
Importance of sensitivity analysis
Benefits of optimal/robust experimental design
Future work
Nonlinear dynamic analysis of limit-cycle oscillation
Sensitivity analysis of oscillatory systems
Acknowledgement
Dr. Martin Brown, Mr. Fei He, Prof. Hong Wang (Control Systems
Centre)
Dr. Niklas Ludtke, Dr. Joshua Knowles, Dr. Steve Wilkinson, Prof.
Douglas B. Kell (Manchester Interdisciplinary Biocentre, MIB)
Prof. David S. Broomhead, Dr. Yunjiao Wang (School of
Mathematics)
Ms. Yisu Jin (Central South University, China)
Mr. Jianfang Jia (Chinese Academy of Sciences)
BBSRC project “Constrained optimization of metabolic and signalling pathway models: towards an understanding of the language of cells
”