Correlation(predicted, true)
advertisement
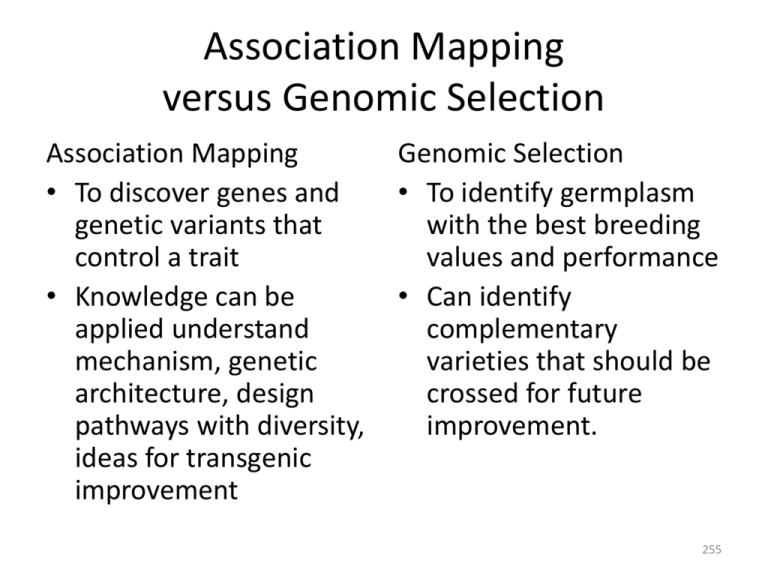
Association Mapping versus Genomic Selection Association Mapping • To discover genes and genetic variants that control a trait • Knowledge can be applied understand mechanism, genetic architecture, design pathways with diversity, ideas for transgenic improvement Genomic Selection • To identify germplasm with the best breeding values and performance • Can identify complementary varieties that should be crossed for future improvement. 255 Association-based selection methods: Genomic selection • We have MAS, why do we need something different? • Historical introduction to genomic selection – – – – – – – The basic idea Methods Theory Selected simulation results Empirical results long-term genomic selection Introgressing diversity using GS 256 MAS problems • Relevant germplasm • Bias of estimated effects • Effects too small for detection 257 Association mapping identifies QTL rapidly while scanning relevant germplasm Intermated recombinant inbreds 5 Research time (year) Positional cloning Near-isogenic lines Recombinant inbred lines Relevance to breeding germplasm High Depends Low Pedigree 1 Association mapping 1 1 x 104 F2 / BC 1 x 107 Resolution (bp) 258 Bias in Effect Estimation Significance Threshold Effect Estimate (True + Error) Average “Detected” Effect Estimated Bias True Effect Locus Effect Estimate • Keep in all loci => No threshold => Estimated effects are unbiased 259 In polygenic traits, much is hidden E.g., h2 = 0.8 α = 0.01 1200 260 Lande & Thompson 1990 Genomic selection principles • Meuwissen et al. 2001 Genetics 157:1819-1829 • No distinction between “significant” and “nonsignificant”; no arbitrary inclusion / exclusion: all markers contribute to prediction • More effects must be estimated than there are phenotypic observations • Estimated effects are unbiased • Capture small effects 261 Genomic selection: Prediction using many markers Breeding Material Genotyping Calculate GEBV Make Selections Meuwissen et al. 2001 Genetics 157:1819-1829 262 Statistical modeling: The two cultures X Observed inputs Nature Can we understand Y? X Regression Observed responses Y Identify causal inputs Y Can we predict Y? X ? Y Regression Decision trees Whatever works Breiman 2001 Stat. Sci. 16:199-231 263 Need to shorten breeding cycle 1 4 3.5 3 2.5 i 2 1.5 1 0.5 0 0.8 0.6 rA 0.4 0.2 0 1 10 100 1000 Ratio Candidates / Selected 10000 1 10 100 Number of Replications 1000 i cumulates over breeding cycles 264 Phenotypic Selection Select Cross Inbreed 1 Season Years F13× Inducer 2 Seasons 1 Rep Self DH0 N=2270 S=100 Phenotype 2 Years 5 Reps N=100 S=10 Release 265 Genomic Selection Select Cross 1 Year! Inbreed Phenotype Release 266 FastGS Select 1 Season = ⅓ Year!! Cross Inbreed Phenotype Release 267 Selection Intensities • Phenotypic – N = 2270, S = 10: i = 2.4 • FastGS (!!!) – N = 370, S = 43: i = 1.7 – 9 × i ≅ 15 Inbreeding: 268 Rates of gain per year 269 Impacts • Schaeffer, L.R. 2006. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 123:218-223. 270 Cost per genetic standard deviation $116 M Genomic Phenotypic Schaeffer 2006 $4.2 M 271 Potential Impact Heffner, E.L. et al. 2009. Genomic Selection for Crop Improvement. Crop Science 49:1-12 Test varieties and release Advance lines informative for model improvement Phenotype (lines have already been genotyped) Model Training Cycle Train prediction model Advance lines with highest GEBV Updated Model Genomic Selection Line Make crosses Development and advance generations Cycle Genotype New Germplasm 272 What (I think) is revolutionary Test varieties and release Advance lines informative for model improvement Phenotype (lines have already been genotyped) Model Training Cycle Train prediction model Advance lines with highest GEBV Updated Model Genomic Phenotypic Selection Selection Line Make crosses Development and advance generations Cycle Genotype New Germplasm For a century, breeding has focused on better ways to evaluate lines. Henceforth it will focus on how to improve a model. 273 A Focus for Information Genomic Prediction Model Development • Current pheno–geno data • Historical pheno–geno data • Linkage and association mapping • Biological knowledge Select Cultivar Release Cross Population Improvement 274 The Alleletarian Revolution • The breeding line as the focus of evaluation has been dethroned in favor of the allele • A line is useful to us only with respect to the alleles it carries • Time-honored practice: replicate (progeny test) lines • But alleles are replicated regardless of what line carries them 275 Methods • Linear models: – Effects are random – Methods differ in marker effect priors • Machine learning methods – Regression trees 276 Linear models: Priors on coefficients • Ridge regression • • BayesB (SSVS) • else • BayesCπ • else 277 Ridge regression BayesB Density BayesCπ Var(β) 278 Machine learning methods • Random Forests – Forest of regression trees – Each tree on a bootstrapped sample – Nodes split on randomly sampled features – Prediction is forest mean 0 0 M2 M1 1 1 0 M2 1 M1 • Can capture interactions M2 0 1 0 1 0 1 0 1 279 Additive models and breeding value • Breeding value = Mean phenotype of progeny – Most important parent selection criterion – Recombination: parents do not always pass combinations of genes to their progeny – > Sum of individual locus effects • Linear models capture this; Machine learning methods may not 280 Theory • How accurate will GS be? • Impact of GS on inbreeding / loss of diversity • Genomic selection captures pedigree relatedness among candidates 281 Prediction accuracy = Correlation(predicted, true) • R = irAσA rA = corr(selection criterion, breeding value) • On simulated data corr(Â, A) is easy • On real data: 282 Predict prediction accuracy • Daetwyler, H.D. et al. 2008. Accuracy of Predicting the Genetic Risk of Disease Using a Genome-Wide Approach. PLoS ONE 3:e3395 • Assume all loci affecting the trait are known and are independent • Assume marker effects are fixed 283 λ 20 10 5 2 1 0.5 0.1 0.02 Replicating hurts: 2000 with 1 plot is better than 1000 with 2 plots 284 Predict prediction accuracy • Hayes, B.J. et al. 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genetics Research 91:47-60. • Detail on the population genetics that drive nG • Assume marker effects are random • Still assume all markers independent and estimated separately 285 Analytical approximations Daetwyler et al., 2008 NP / N G Hayes et al., 2009 NP / N G 286 Take Homes • Even with traits of very low heritability (h2 = 0.01), sufficient nP gives accuracy • Replication may not be good • The number of loci estimated (nG) is a critical parameter • If you don’t know where the QTL are, higher marker coverage requires higher nG • N.B. All conclusions assuming only 100% LD! 287 Genetic diversity loss / inbreeding • Daetwyler, H.D. et al. 2007. Inbreeding in genome-wide selection. J. Anim. Breed. Genet. 124:369-376 • Avoid selecting close relatives together • What is the correlation in the estimated breeding value between full sibs? Correlation sibling estimates 288 Genetic diversity loss / inbreeding _BLUP_ Mendelian sampling term σ2B Aj = ½AS + ½AD + aj σ2W = 0 __GS__ σ2B Correlation sibling estimates σ2W > 0 289 Daetwyler et al. 2007 Take Homes • Genomic selection captures the Mendelian sampling term. – Correlation between the estimates of sibling performance are reduced – Co-selection of sibs is reduced – Rate of inbreeding / loss of diversity is reduced 290 A word on pedigree relatedness • Five individuals, a, b, c, d, and e. – a, b, and c unrelated – d offspring of a and b – e offspring of a and c A= a b a 1 0 b 0 1 c 0 0 d ½ ½ e ½ 0 c d 0 ½ 0 ½ 1 0 0 1 ½ ¼ e ½ 0 ½ ¼ 1 291 Ridge Regression Habier, D. et al. 2007. Genetics 177:2389-2397 Hayes, B.J. et al. 2009. Genetics Research 91:47-60. 292 Habier et al. simulation set up 293 Genetic relationship decays fast • Prediction from pedigree relationship loses acccuracy very quickly • Decay rate is initially more rapid then stabilizes after about 5 generations • Rapid initial decay reflects that the closest marker may not be in highest LD with the QTL • RR-BLUP accuracy decays more rapidly than Bayes-B because more markers absorb the effect of a QTL Training population here 294 Habier et al. 2007 Take homes • The ability of genomic selection to capture information on genetic relatedness is valuable • That information decays rapidly • The amount of that information relates to the number of markers fitted by a model: – Ridge regression > BayesB • Bayes-B captured more LD information: – Long-term accuracy: BayesB > Ridge regression 295 Accuracy due to relationships vs. LD 296 Stochastic vs deterministic prediction Habier et al. Zhong et al. NP / N G 297 To replicate or not to replicate 504 Lines replicated once Ridge Regression 168 Lines replicated three times BayesB 298 Genetic diversity loss / inbreeding _BLUP_ Mendelian sampling term σ2B Aj = ½AS + ½AD + aj σ2W = 0 Correlation sibling estimates __GS__ σ2B σ2W > 0 Capturing relationship Information increases σ2B NOT σ2W 299 Simulation setting: Meuwissen; Habier; Solberg • Ne = 100; 1000 generations • Mutation / Drift / Recombination equilibrium • High marker mutation rate (2.5 x 10-3 / loc / gen); higher “haplotype mutation rate” • Mutation effect distribution Gamma (1.66, 0.4): “effective QTL number” is only about 6 (!) – > Watch out how you simulate! 300 Results • Prediction accuracy estimated by simulation MHG HFD RR-BLUP 0.73 0.64 BayesB 0.85 0.69 • These accuracies are ASTOUNDING • If h2 = 1, r = 0.71 301 Noteworthy discussion • Markers flanking QTL not always in model – QTL effects captured by multiple markers – No need to “detect” QTL • Recombination causes accuracy to decay – Faster than if QTL captured by flanking markers – Markers far from QTL contribute to capture its effect • Ne / 2 markers per Morgan achieves close to maximum accuracy – Dependent on high marker mutation rates (?) 302 Solberg et al. 2008 • Density: Number of markers per Morgan SNP: SSR: ¼ Ne 1 Ne ½ Ne 2 Ne 1 Ne 4 Ne 2 Ne 8 Ne 303 Zhong et al. 2009 • Zhong, S. et al. 2009. Genetics 182:355-364. • 42 diverse 2-row barley • 1040 markers ~ evenly spaced • Mating designs to generate 500 high and low LD training dataset • 20 or 80 QTL; h2 = 0.4 304 Ridge regression Vs. BayesB QTL: Observed Unobserved 20QTL – HiLD 20QTL – LoLD Ridge Regression 80QTL – HiLD 80QTL – LoLD BayesB 305 Zhong et al. 2009 Take-home messages • Ridge regression is not affected by the number of QTL / the QTL effect size • BayesB performs better with large markerassociated effects • Co-linearity is more detrimental to BayesB • High marker density and training pop. size? Yes: BayesB No: RR-BLUP 306 VanRaden et al. 2009 • VanRaden, P.M. et al. 2009. Invited Review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 92:16-24. 307 VanRaden et al. 2009 • Some traits have major genes, others do not 308 VanRaden et al. 2009 • The larger the training population, the better. Where diminishing returns will begin is not in sight. Predictor 309 Take Homes • • • • Training population requirements very large BayesB did not help == no large marker-associated effects == Like the “Case of the missing heritability” in human GWAS studies – Are many quantitative traits driven by very low frequency variants? – RR would capture this case better than BayesB 310 Empirical data on crops: TP size 311 Empirical data on crops: Marker No. 312 Empirical data on Humans: Marker No. Out of 295K SNP Yang et al. 2010. Nat. Genet. 10.1038/ng.608 313 Long-term genomic selection • • • • • Marker data from elite six-row barley program 880 Markers 100 hidden as additive-effect QTL Evaluate 200 progeny, select 20 Phenotypic compared to genomic selection 314 Breeding / model update cycles Season 1 Season 2 Season 3 Season 4 Season 5 Season 6 Cross & Inbreed Evaluate & Select Cross & Inbreed Evaluate & Select Phenotypic Selection Cross & Inbreed Evaluate & Select Genomic Selection Cross & Inbreed Evaluate & Select Evaluate Cross, Inb. & Select Cross, Inb. & Select Evaluate Cross, Inb. & Select Cross, Inb. & Select Evaluation is possible every other season. Candidates from every other cycle can be evaluated. There is still a lag: Parents of C2 are selected based on evaluation of C0. 315 Mean Genotypic Value Response in genotypic value Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle 316 Mean Realized Accuracy Accuracy Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle 317 Mean Genotypic Standard Deviation Genetic variance Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle 318 Mean Number Lost Favorable Allleles Lost favorable alleles Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle 319 Goddard 2008; Hayes et al. 2009 320 Response in genotypic value Weighted Mean Genotypic Value Unweighted Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle Phenotypic Breeding Cycle 321 Genetic variance Mean Genotypic Standard Deviation Unweighted Weighted Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle Phenotypic Breeding Cycle 322 Lost favorable alleles Mean Number Lost Favorable Alleles Unweighted Weighted Phenotypic Selection Genomic; Small Training Pop Genomic; Large Training Pop Phenotypic Breeding Cycle Phenotypic Breeding Cycle 323 Long term genomic selection • The acceleration of the breeding cycle is key • Some favorable alleles will be lost – Likely those not in LD with any marker • Managing diversity / favorable alleles appears a good idea • This can be done using the same data as used for genomic prediction 324 Introgressing diversity • GS relies on marker–QTL allele association • An “exotic” line comes from a sub-population divergent from the breeding population • After sub-populations separate – Drift moves allele frequencies independently – Drift & recombination shift associations independently • Will the GS prediction model identify valuable segments from the exotic? 325 Three approaches • Create a bi-parental family with the exotic (Bernardo 2009) – Develop a mini-training population for that family – Improve the family – Bring it into the main breeding population • Develop a separate training population for the exotic sub-population (Ødegård et al. 2009) • Develop a single multi-subpopulation (specieswide?) training population (Goddard 2006) 326 Need higher marker density Ancestral LD sub-population specific LD • Tightly–linked: ancestral LD • Loosely–linked: sub-population specific LD 327 Consistency of association across barley subpopulations 1.0 0 cM recombination distance 5 cM recombination distance Correlation of r 0.8 0.6 0.4 0.2 0.0 0.0 0.5 Genetic Distance 328 Example: Dairy cattle breeds 0.7 VP = Holstein Prediction Accuracy 0.6 VP = Jersey 0.5 0.4 0.3 0.2 0.1 0 TP = Hols. TP = Jers. Hols. + Jers. 329 Oat sub-populations (UOPN) G1 G2 G3 N=136 N=149 N=161 330 Combined sub-population TP (β-Glucan) 0.11 VP TP G1 G2 and G3 0.50 G1 and G2 0.39 G3 G1 G2 G3 331 Introgressing diversity using GS • Need higher marker density • Analysis of consistency of r may indicate whether current density is sufficient – Not sure we have it for barley • If you have the density, a multi-subpopulation training population seems like a good idea – Focuses the model on tighter ancestral LD rather than looser sub-population specific LD 332








