Catalytic Mechanism of Chymotrypsin
advertisement

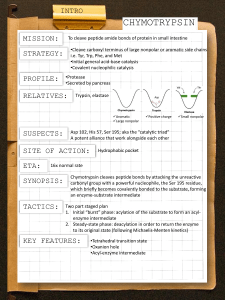
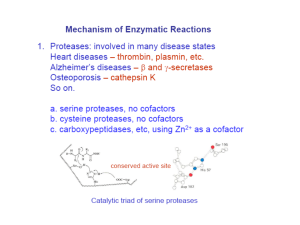
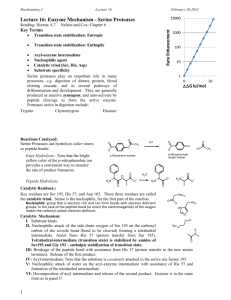
Catalytic Mechanism of Chymotrypsin slide 1 • Chymotrypsin – Protease: catalyze hydrolysis of proteins in small intestine – Specificity: Peptide bond on carboxyl side of aromatic side chains (Y, W, F) & Large hydrophobic residues (Met,…) – Three polypeptide chains cross-linked to each other – Three catalytic residues: Ser195, His57, & Asp102 Catalytic Mechanism of Chymotrypsin slide 2 Catalytic Mechanism of Chymotrypsin slide 2 Catalytic Mechanism of Chymotrypsin slide 3 Catalytic Mechanism of Chymotrypsin slide 4 Summary for the Catalytic Mechanism of Chymotrypsin • Mechanism – General acid-base catalysis & Covalent catalysis – Two steps: Acylation & Deacylation (rate limiting; reverse of acylation with water substituting the amine component) – Key features • Active Ser195 & roles of the three catalytic residues • Tetrahedral transition state • Oxyanion and Oxyanion hole • Acyl-enzyme intermediate Serine Protease Family Chymotrypsin & elastase main chain conformation (superimposed) • Serine Proteases – Chymotrypsin – Trypsin – Elastase • Similarity – Similar 3D structure – Catalytic triad – Oxyanion hole – Covalent acyl-enzyme intermediate – Secreted by pancrease as inactive precursors Specificity Difference of Chymotrypsin, Trypsin, and Elastase • Substrate specificity – Chymotrypsin: aromatic or bulky nonpolar side chain – Trypsin: Lys or Arg – Elastase: smaller & uncharged side chains • Small structural difference in the binding site explains the substrate specificity nonpolar pocket Asp (negatively charged) vs. Ser in Chymotrypsin no pocket present as two Gly in chymotrypsin are replaced by Val and Thr Carboxypeptidase A • Digestive enzyme • Hydrolyzes carboxyl terminal peptide bond – Prefer bulky and aliphatic residues • 3D structure – Single polypeptide (307 amino acids) – helices (38%) and (17%) (compact, ellipsoid) A tightly bound Zn2+ Essential for catalysis Coordinated to 1 H2O, 2 His, 1 Glu Substrate Binding Induces Large Structural Changes at the Active Site Substrate Binding Induces Large Structural Changes at the Active Site • 3D Structure of peptidase A/glycyltyrosine complex – Substrate-induced structural change at active site • 12 Å movement of Tyr248-OH & rotation (Moves from surface to substrate terminal COO-) – New interaction: Tyr248 OH –OC=O – Closes active-site cavity – Extrude water from cavity • Arg145 moves 2 Å – New interaction: Arg145 & –OC=O (substrate) • Terminal side chain of substrate – Now sits in a hydrophobic pocket – Induced-fit model (Daniel Koshland, Jr.) Substrate Binding at the Active Site Catalytic Mechanism of Carboxypeptidase A • The H2O molecule is activated by – Bound Zn2+ and COO– of Glu270 • Activated H2O attacks the C=O group of the scissile peptide bond • Glu270 simultaneously accepts a H+ from H2O • A negatively charged tetrahedral intermediate is formed • Intermediate is stabilized by Zn2+ and Arg127 • H+ transfer from COOH of Glu270 to the peptide NH • Peptide bond is concomitantly cleaved • The reaction products diffuse away • Summary: – Activation of H2O by Zn2+ and Glu270 – Proton abstraction and donation by Glu270 – Electrostatic stabilization of tetrahedral intermediate by Arg127 and Zn2+ Catalytic Mechanism of Carboxypeptidase A